- Title
-
Pax6 organizes the anterior eye segment by guiding two distinct neural crest waves
- Authors
- Takamiya, M., Stegmaier, J., Kobitski, A.Y., Schott, B., Weger, B.D., Margariti, D., Cereceda Delgado, A.R., Gourain, V., Scherr, T., Yang, L., Sorge, S., Otte, J.C., Hartmann, V., van Wezel, J., Stotzka, R., Reinhard, T., Schlunck, G., Dickmeis, T., Rastegar, S., Mikut, R., Nienhaus, G.U., Strähle, U.
- Source
- Full text @ PLoS Genet.
|
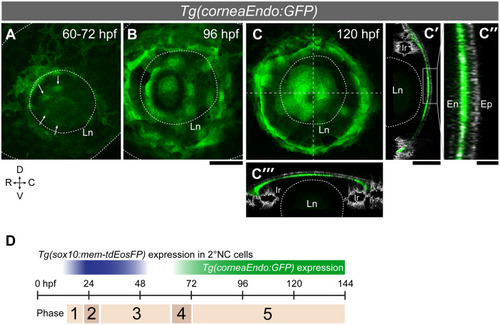
(A-J”) Confocal fluorescent images of |
|
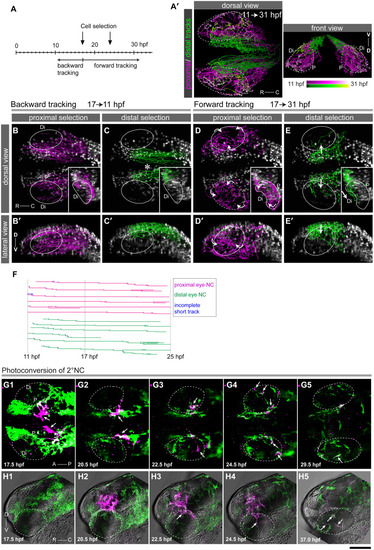
Dorsal views of a |
|
DSLM time series of a double transgenic embryo expressing nuclear Eos in NC cells [green, |
|
(A-C”) Double EXPRESSION / LABELING:
|
|
(A-C”) The |
|
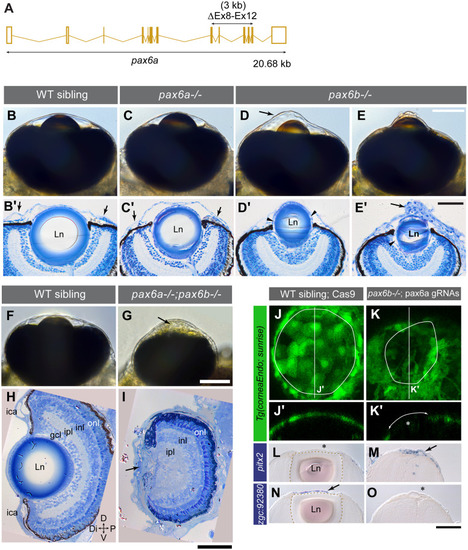
(A) CRISPR/Cas9-mediated deletion of 3 kb in the |
|
|
|
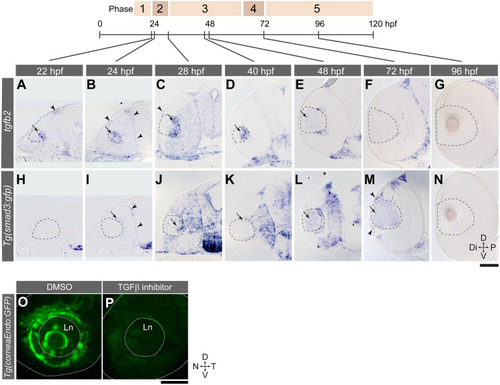
Gene expression was analysed by EXPRESSION / LABELING:
PHENOTYPE:
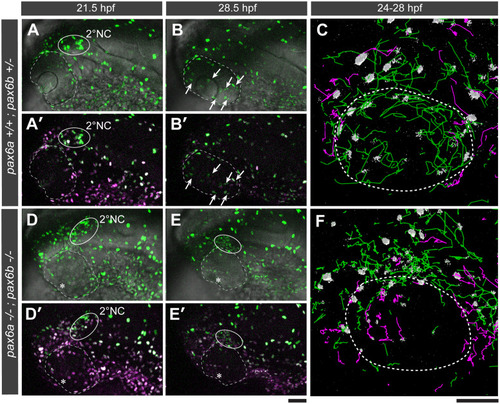
|
|
Intravenous injection of 2000 kDa dextran (green) and 3 kDa dextran (magenta) into wild type (A-B) and double homozygous |
|
(A-N) |