- Title
-
Tissue-Specific Requirement for the GINS Complex During Zebrafish Development
- Authors
- Varga, M., Csályi, K., Bertyák, I., Menyhárd, D.K., Poole, R.J., Cerveny, K.L., Kövesdi, D., Barátki, B., Rouse, H., Vad, Z., Hawkins, T.A., Stickney, H.L., Cavodeassi, F., Schwarz, Q., Young, R.M., Wilson, S.W.
- Source
- Full text @ Front Cell Dev Biol

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
|
|
PHENOTYPE:
|
|
|
|
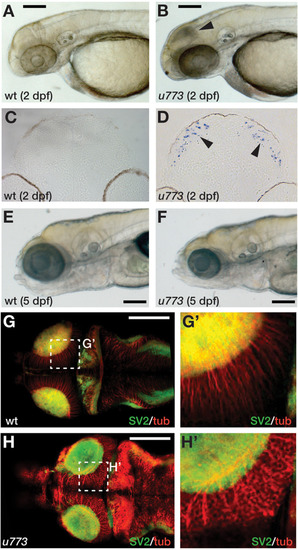
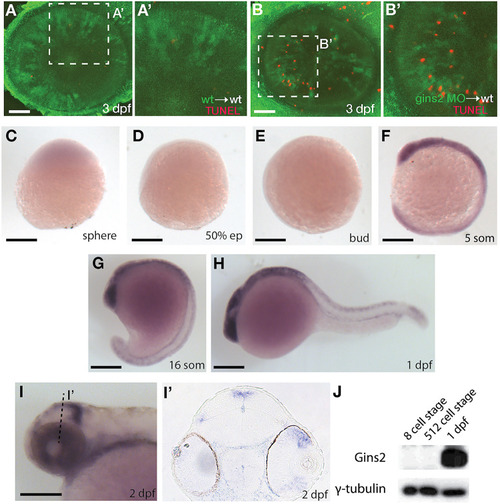
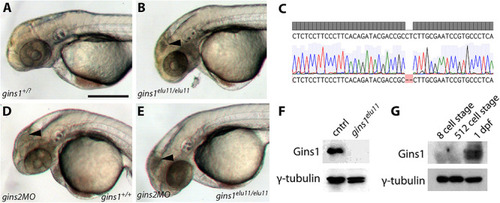
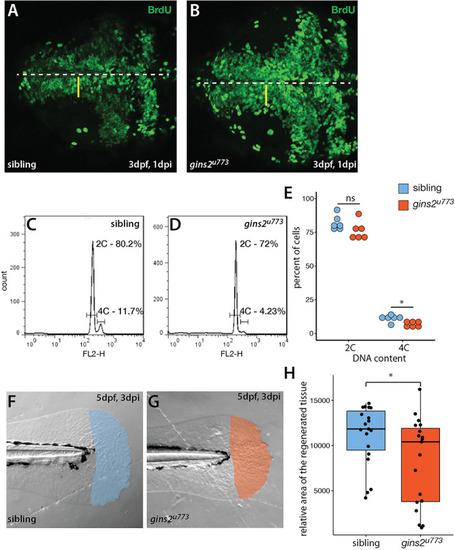
Gins1 deficient and Gins1, Gins2 double-deficient embryos show comparable cell death phenotypes in eyes and tecta. |
|
|
|
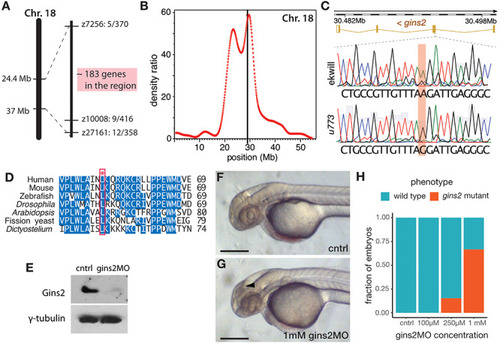
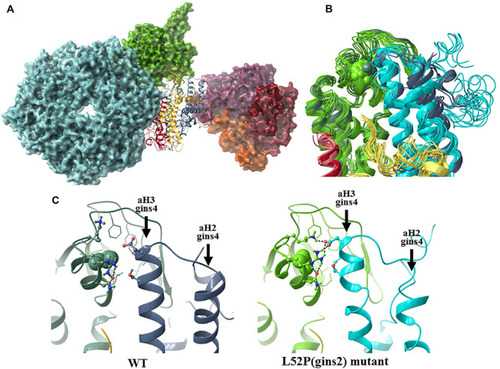
The structural consequences of the Gins2L52P mutation in the zebrafish GINS complex. |