- Title
-
Gon4l/Udu Regulates Cardiomyocyte Proliferation and Maintenance of Ventricular Chamber Identity During Zebrafish Development
- Authors
- Budine, T.E., de Sena-Tomás, C., Williams, M.L.K., Sepich, D.S., Targoff, K.L., Solnica-Kreze, L.
- Source
- Full text @ Dev. Biol.
|
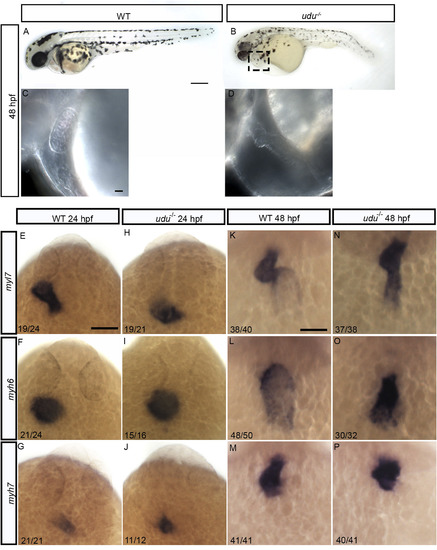
Loss of zygotic Gon4l function affects cardiac chamber patterning and causes a reduction in number of cardiomyocytes. Lateral view of WT embryos (A) and udu−/− (B) and their heart region (C, D) at 48 hpf. Dorsal view of WT (E, F, G) and udu−/- embryos (H, I,J) showing the expression of myl7 (E, H), myh6 (F, I), and myh7 (G, J) at 24 hpf by WISH. Dorsal view of myl7 (K, N), myh6 (L, O), and myh7 (M, P) expression at 48 hpf by WISH in WT (K, L, M) udu−/− (N, O, P) embryos. |
|
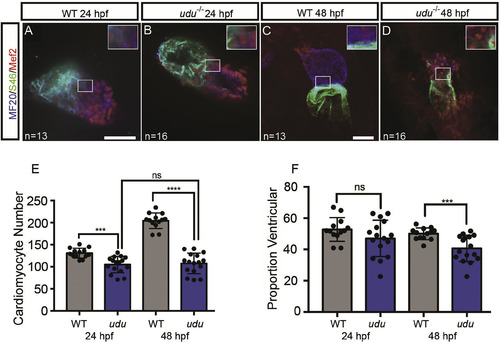
Loss of zygotic Gon4l function affects cardiac chamber patterning after heart tube formation and results in a reduction in the number of cardiomyocytes. Immunofluorescence confocal images of cardiac chambers at 24 hpf using MF20 (blue, myocardium) and S46 (green, atrium) and Mef2 (red, cardiomyocyte nuclei) antibodies to mark the cardiac chambers in WT (A, C) and udu−/− mutant (B, D) embryos at 24 hpf (A, B) and 48 hpf (C, D). Cardiomyocyte number (E) and percentage of total cardiomyocytes with ventricular identity (F) at 24 hpf and 48 hpf in WT (grey) and udu−/− mutant (blue) embryos. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, error bars = SEM. Scale bars represent 50 μm. Inserts show a 2X magnification from images. |
|
EdU incorporation is reduced in the hearts of udu−/− embryos. Experimental workflow for EdU labeling (A). EdU labeling (red) in 48 hpf WT hearts (B) and udu−/− hearts (C) with cardiomyocytes labeled by Mef2 (blue). Arrows showing EdU + cardiomyocyte (white) and non-cardiomyocyte (green). Graph showing the proliferative index for EdU in WT (grey) and udu−/−(blue) cardiomyocytes (D). qRT-PCR for ccnd1 (E), ccnd2a (F), ccnd2b (G), ccnd3 (H), ccne1 (I), and ccne2 (J) performed on RNA extracted from hearts isolated from 36 hpf WT (grey) and udu−/−(blue) embryos. Results are shown as fold change of gene expression in udu−/− embryos compared to WT and results were standardized to gapdh expression. DamID-seq Genome browser tracks of Gon4l-Dam and GFP Control-Dam at ccnd1 (E′), ccnd2a (F′), ccnd2b (G′), ccnd3 (H′), ccne1 (I′), and ccne2 (J′) loci. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, error bars = SEM. Scale bar represents 50 μm. |
|
Expression of nkx2.5 and nkx2.7 is initiated normally in the nascent cardiomyocytes of udu−/− embryos, but becomes reduced following heart tube formation. Dorsal view of embryos showing expression of nkx2.5 (A–F) and nkx2.7 (G–L) detected by WISH in WT (A, C, E, G, I, K) and udu−/− (B, D, F, H, J, L) embryos at 12 somites (A, B, G, H), 16 somites (C, D, I, J) and 24 hpf (E, F, K, L) stages. DamID-seq Genome browser tracks of Gon4l-Dam and GFP Control-Dam at nkx2.5 (M) and nkx2.7 (N) loci. Graph of qRT-PCR results performed on RNA extracted from hearts isolated from 36 hpf and 48 hpf WT and udu−/− embryos for nkx2.5 (O) and nkx2.7 (P) expression; results were standardized to gapdh expression. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, error bars = SEM. Scale bars represent 50 μm. |
|
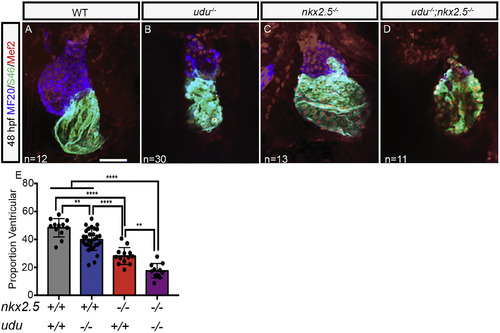
udu genetically interacts with nkx2.5 to maintain ventricular identity. Confocal immunofluorescence images of ventral views of WT (A), udu−/− (B), nkx2.5−/− (C), and udu−/−; nkx2.5−/−(D) zebrafish hearts at 48 hpf with showing the myocardium (MF20, blue) and atrium, (S46, green), and nuclei of cardiomyocytes (Mef2, red). Proportion of total cardiomyocytes with ventricular identity at 48 hpf in WT (grey), udu−/−;nkx2.5+/+ (blue) udu+/+;nkx2.5−/− (red) and udu−/−;nkx2.5−/− (purple) embryos (E). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, error bars = SEM. Scale bars represent 50 μm. |
|
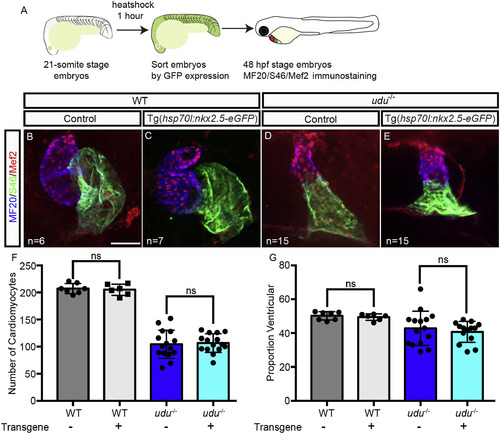
Ectopic nkx2.5 expression is not sufficient to rescue ventricular identity in udu−/− embryos. Experimental workflow (A). Immunofluorescent labeling with S46 (green) labeling the atrium, MF20 (blue) labeling the myocardium and Mef2 (red) labeling the cardiomyocyte nuclei in 48 hpf WT embryos (B, C) and udu−/− embryos without Tg(hsp70l:nkx2.5-eGFP) (B, D) and those harboring Tg(hsp70l:nkx2.5-eGFP) (C, E). Graphs showing the total number of cardiomyocytes (F), and a proportion of ventricular cardiomyocytes (G) in WT (grey) and udu−/− (blue) embryos with (light grey, light blue) and without Tg(hsp70l:nkx2.5-eGFP) (dark grey, dark blue). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, error bars = SEM. Scale bar represents 50 μm. |
Reprinted from Developmental Biology, 462(2), Budine, T.E., de Sena-Tomás, C., Williams, M.L.K., Sepich, D.S., Targoff, K.L., Solnica-Kreze, L., Gon4l/Udu Regulates Cardiomyocyte Proliferation and Maintenance of Ventricular Chamber Identity During Zebrafish Development, 223-234, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.