- Title
-
Oligodendrocytes express synaptic proteins that modulate myelin sheath formation
- Authors
- Hughes, A.N., Appel, B.
- Source
- Full text @ Nat. Commun.
|
Axons accumulate synaptic vesicle release machinery under myelin sheaths. EXPRESSION / LABELING:
|
|
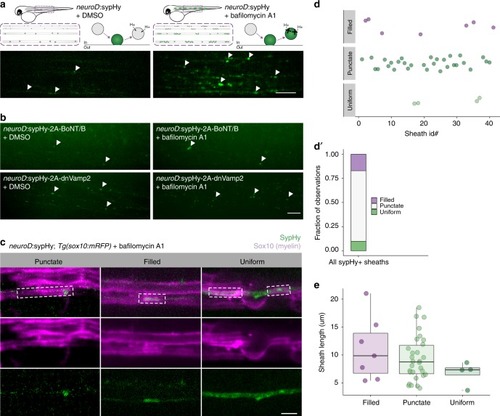
Variable synaptic vesicle exocytosis sites under myelin sheaths. |
|
PSD95 is expressed by myelinating oligodendrocytes and is variably localized within myelin sheaths. EXPRESSION / LABELING:
|
|
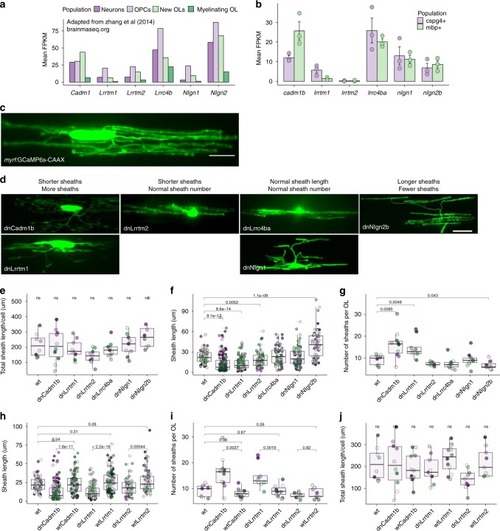
Candidate synaptogenic adhesion molecules have variable effects on myelin sheath length and number. |
|
Cadm1b localizes to myelin sheath membrane. EXPRESSION / LABELING:
|
|
The extracellular, trans-acting adhesion domain (Ig1) of Cadm1b promotes myelin sheath growth. |