- Title
-
β-glucan-dependent shuttling of conidia from neutrophils to macrophages occurs during fungal infection establishment
- Authors
- Pazhakh, V., Ellett, F., Croker, B.A., O'Donnell, J.A., Pase, L., Schulze, K.E., Greulich, R.S., Gupta, A., Reyes-Aldasoro, C.C., Andrianopoulos, A., Lieschke, G.J.
- Source
- Full text @ PLoS Biol.
|
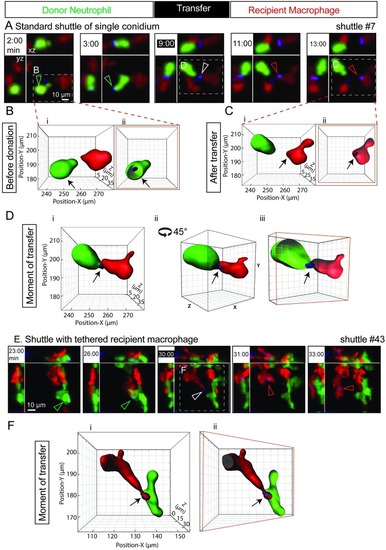
(A) A representative standard shuttle of a calcofluor-stained conidium (blue) from a |
|
A variety of shuttles of conidia (blue) from |
|
(A) Shuttle of calcofluor-stained conidium (blue) from |
|
(A,B) Relative frequency of shuttles for different cargos, incidence computed for each condition as number of 3-h imaging datasets with shuttle(s)/total number of imaging datasets. By chi-squared analysis, there are no significant differences for the comparisons: live spores of the two species ( |
|
(A,B) Two sequences demonstrating neutrophil-to-macrophage shuttling of Alexa-Fluor-488–labeled zymosan particle between murine phagocytes in vitro. Panel (i) is a schematic showing the elongated, adherent recipient macrophage. Panels (ii–viii) are bright-field photomicrographs with green fluorescence channel overlaid, with time points indicated in min:s. Red arrow indicates the shuttled particle in donor neutrophil (panels ii–vi) and then, following shuttling, within the recipient macrophage (panels vii–viii). Stills from |
|
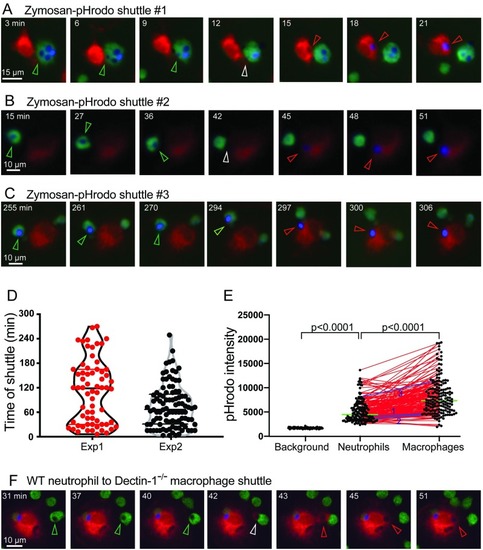
(A–C) Three examples of in vitro shuttling of zymosan–pHrodo particles, representative of 164 shuttling events. Neutrophils, green; macrophages, red; pHrodo–zymosan, false-colored blue. Scales as shown. Stills from |