- Title
-
On the traces of tcf12: Investigation of the gene expression pattern during development and cranial suture patterning in zebrafish (Danio rerio)
- Authors
- Blümel, R., Zink, M., Klopocki, E., Liedtke, D.
- Source
- Full text @ PLoS One
|
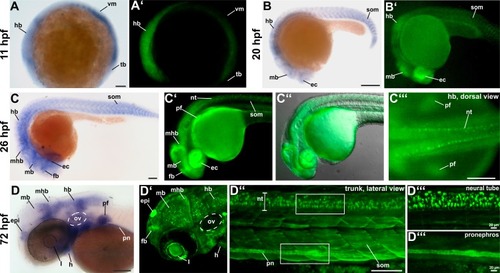
(A, A’) EXPRESSION / LABELING:
|
|
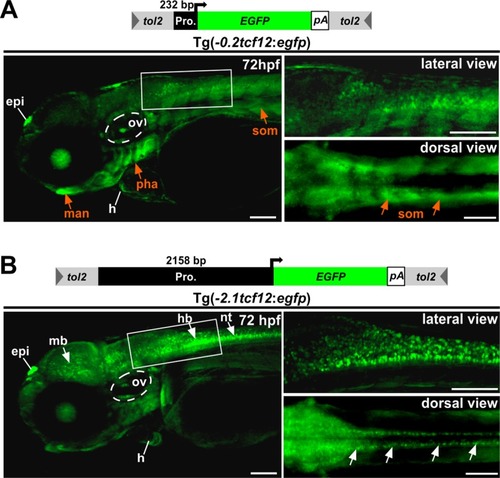
Comparison between different EXPRESSION / LABELING:
|
|
Investigation of EGFP expression over time |
|
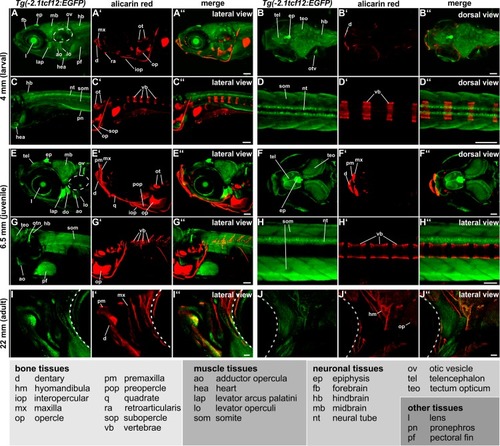
Dorsal views of 8–18 mm (30–90 dpf) zebrafish skull vaults are depicted. Mineralized structures were stained with alizarin red. The schemes illustrate the growth stage of the calvarial bones. Red squares enclose the regions of the calvaria that are shown on the confocal images. EGFP signals in the center of the developing skull plates in A-E derive from the overlaying skin (marked with asterisk). C’ and E’ show detail magnifications of the boxed regions in C and E. The images display maximum intensity Z-projections from confocal stacks. Dashed white lines in I indicate fronts of the overlapping frontal and parietal bones. a, anterior; c, coronal suture; fb, frontal bone; if, interfrontal suture; l, lambdoid suture; p, posterior; pb, parietal bone; s, sagittal suture; sop, supraoccipital. All scale bars represent 100 μm. EXPRESSION / LABELING:
|
|
Confocal images of immunostaining on cryosections of the frontal and parietal bones and the coronal sutures of adult EXPRESSION / LABELING:
|
|
(A) Graphic representation of the location of EXPRESSION / LABELING:
|
|
Suture visualization in zebrafish.Alizarin red staining was performed at different stages of development (A: 15 mm, 50 dpf; B: 26 mm, 120 dpf; C: 36 mm, 280 dpf/adult) and showed calvarial plate growth and progression of suture establishment over time. Dorsal views are maximum intensity Z-projections from confocal stacks. |