- Title
-
Disruption of asxl1 results in myeloproliferative neoplasms in zebrafish
- Authors
- Gjini, E., Jing, C.B., Nguyen, A.T., Reyon, D., Gans, E., Kesarsing, M., Peterson, J., Pozdnyakova, O., Rodig, S.J., Mansour, M.R., Joung, K., Look, A.T.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
PHENOTYPE:
|
|
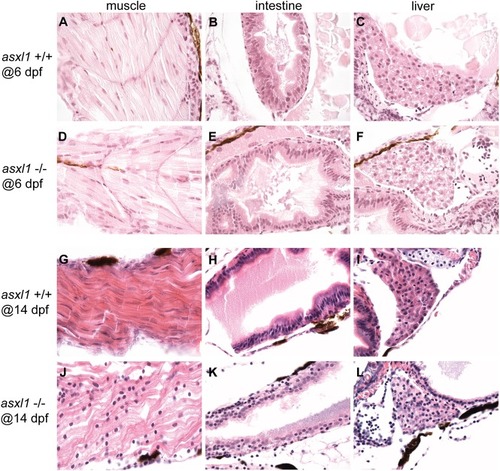
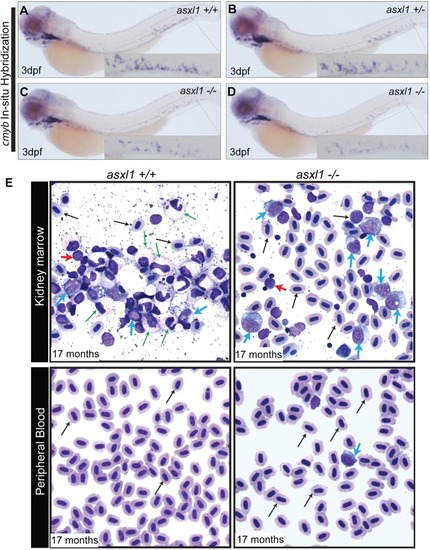
Histopathological analysis of kidney, intestine and liver development in 17-month-old asxl1+/+ and asxl1+/- and asxl1-/- fish. Results revealed that intestine (D, E and F) and liver (G, H and I) was normal and similar to that of the fish, indicating that these fish recovered from early developmental hypoplasia in these organs. However, asxl1-/- mutants did have decreased numbers of erythroid islands in the kidney marrow compared with asxl1+/+ and asxl1+/- fish (A, B and C), indicating that the hematopoietic system is abnormal in these animals. Area encircled with black line indicates erythroid islands. The quantification of erythroid islands was shown in (J), with the black bars representing the median values. ns p>0.05, *p<0.05, **p<0.01 by two-tailed unpaired t-test. PHENOTYPE:
|
|
Whole-mount in situ hybridization (WISH) for gata-1 (22 hours-post-fertilization), l-plastin and mpx (22 hours-post-fertilization) was performed in asxl1+/+, asxl1+/-, asxl1-/-, asxl1+/+tet2 -/-, asxl1+/-tet2 -/- and asxl1-/-tet2 -/- zebrafish embryos. This assay shows that there is no difference in the number of cells expressing gata-1, l-plastin and mpx among all the different genotypes. |
|
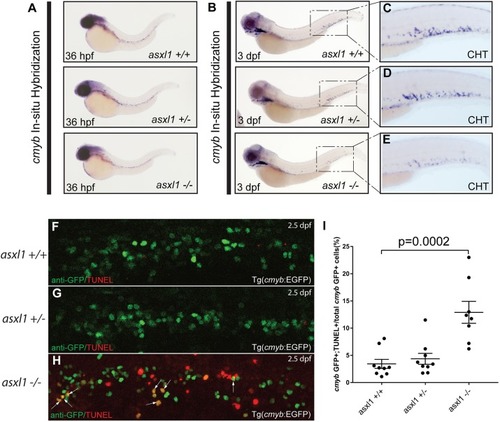
The number of HSPCs in the CHT is reduced in the asxl1 -/- embryos at 3 dpf. (A-C) EGFP+ HSPCs in asxl1 +/-, asxl1 +/- and asxl1 -/- embryos. (D) Statistical analysis of HSPC numbers in asxl1 +/-, asxl1 +/- and asxl1 -/- embryos indicate that EGFP+ HSPCs are decreased in CHT region in asxl1 -/- embryos at 3 dpf |
|
EGFP+ HSPCs in asxl1 +/+, asxl1 +/- and asxl1 -/- embryos at 3 dpf. Knockdown of bim (D-F) and bid (G-I) or overexpression of bcl2 (J-L) each were able to partially rescue HSPC numbers in asxl1 -/- embryos compared with control embryos. This result is expected because mitochondrial intrinsic programmed cell death depends on the combined activities of BH3-only pro-apoptotic proteins and is blocked by prosurvival proteins like bcl2. PHENOTYPE:
|