- Title
-
A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates
- Authors
- Parker, H.J., De Kumar, B., Green, S.A., Prummel, K.D., Hess, C., Kaufman, C.K., Mosimann, C., Wiedemann, L.M., Bronner, M.E., Krumlauf, R.
- Source
- Full text @ Nat. Commun.
|
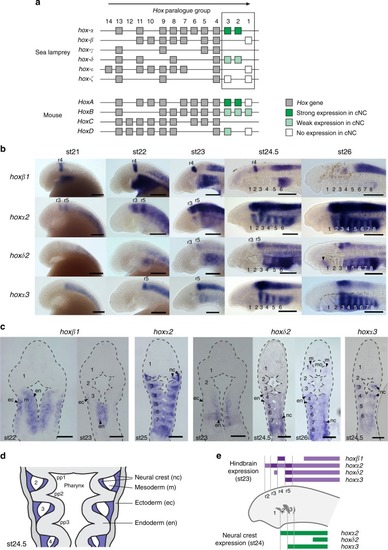
Embryonic time course showing expression of |
|
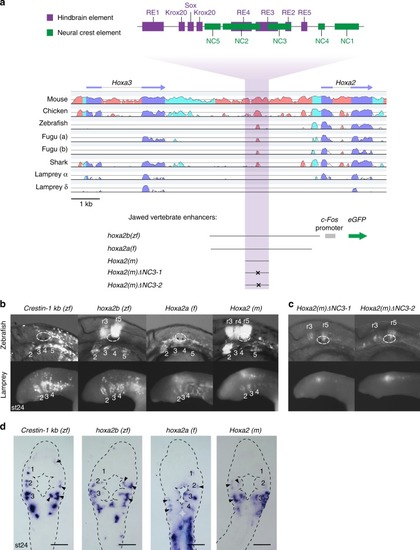
Conserved activity of gnathostome |
|
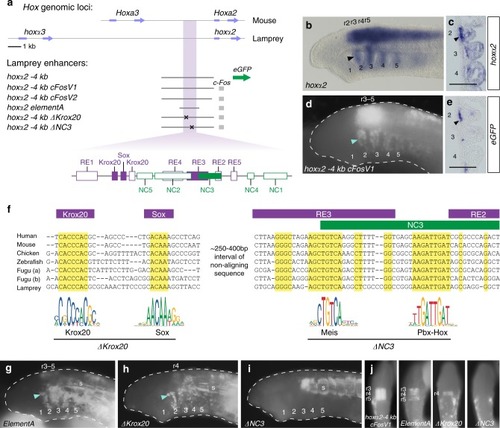
Characterization of a lamprey |
|
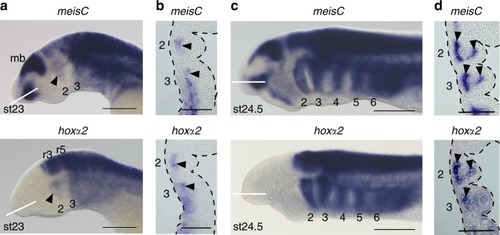
Endogenous expression of |
|
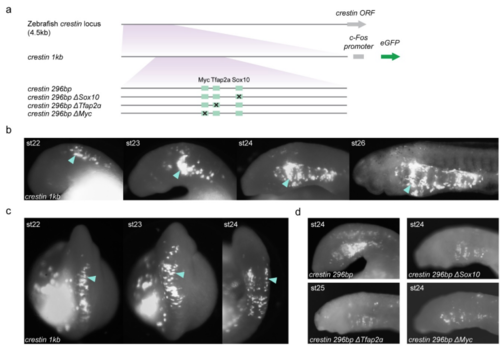
GFP reporter expression in NC mediated by variants of the zebrafish crestin promoter/enhancer in transient transgenic lamprey embryos. a, Versions of the zebrafish crestin promoter/enhancer tested for activity in zebrafish and lamprey embryos in this study. In zebrafish, the NC-specific activity of the crestin element depends upon consensus transcription factor binding sites for multiple transcription factors that are known to be part of a core NCGRN, including Sox10, Tfap2α and cMyc. Variants of the core minimal promoter/enhancer (crestin 296bp) were generated with mutations in these sites(ΔSox10, ΔTfap2α, ΔMyc). Regulatory elements were cloned upstream of the mouse c-Fos promoter. b-c, Lateral (b) and dorsal (c) views of two different transient transgenic lamprey embryos exhibiting GFP expression in NC (arrowheads) under the control of the crestin 1kb promoter/enhancer. Expression is first seen in the dorsal neural tube in NC cells as they start to delaminate, which then migrate to populate the pharyngeal arches at later stages. Even though the crestin element is specific to zebrafish and is not present in other gnathostomes, its cis-regulatory activity is conserved between lamprey and zebrafish. These data suggest that upstream regulatory factors that mediate activity of the crestin element in zebrafish may also be present in lamprey NC. d, Lateral views of st24 lamprey embryos injected with variants of the crestin promoter/enhancer. A minimal crestin promoter/enhancer is active in the lamprey NC; we found that the activity of this element in lamprey is also sensitive to perturbation of the same binding sites critical for activity in zebrafish. These results, coupled with the endogenous expression of SoxE1-3, Tfap2, and n-Myc in lamprey, suggest that the crestin element is interpreted in lamprey by components of an ancestral NC GRN that includes Sox, Tfap2α and Myc factors. GFP-expressing embryos shown are representative of the expression potential of the reporter construct in each case, as inferred from screening many (typically more than 100) injected embryos. Supplementary Tables 1-2 provide the injection statistics for crestin constructs in zebrafish and lamprey embryos. |
|
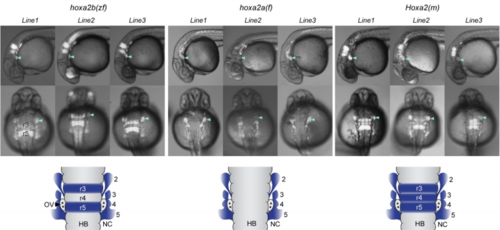
GFP reporter expression driven by gnathostome Hoxa2 NC enhancers in transgenic zebrafish reporter lines. Three independent lines are shown for each of the three gnathostome Hoxa2 enhancers. Letters in parenthesis indicate species of origin of the enhancer: zf, zebrafish; f, fugu; m, mouse. Lateral (top) and dorsal (middle) views are shown. Arrowheads denote GFP expression in PA2. Schematics at the bottom depict dorsal views of the rhombomeres (r) and pharyngeal arches (numbered), illustrating the consistent domains of activity observed between separate transgenic lines for each enhancer (blue shading). Abbreviations: HB, hindbrain; NC, neural crest; OV, otic vesicle. |
|
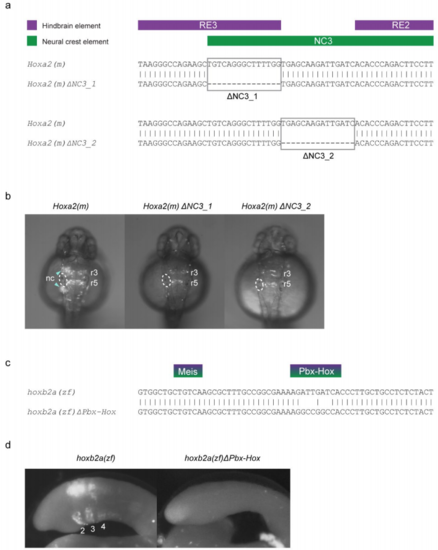
Mutation of sites within gnathostome Hox2 enhancers and their influence on tissue-specific activity in transient transgenic zebrafish and lamprey embryos. a, Alignments of the NC3 region of the wild-type mouse (m) Hoxa2 NC enhancer with two variants, ΔNC3_1 and ΔNC3_2, showing the 15bp portions deleted in each variant. The positions of characterised hindbrain (purple) and NC (green) cis-elements are shown above the alignments. b, Dorsal views of transient transgenic zebrafish embryos with GFP expression mediated by the wild-type Hoxa2(m) enhancer and the two ΔNC3 variants. The left otic vesicle of each embryo is circled, with GFP expression in rhombomeres (r) and neural crest (nc) annotated. Embryos are at approximately 30 hours post-fertilisation. c, Alignment of a portion of the wild-type zebrafish (zf) hoxb2a NC enhancer with a variant in which the Pbx-Hox site has been mutated (ΔPbx-Hox). d, st24 transient transgenic lamprey embryos injected with the wild-type hoxb2a(zf) (lateral view) and mutated hoxb2a(zf)ΔPbx-Hox (dorsal view) enhancers. hoxb2a(zf) mediates expression in the hindbrain and neural crest, posterior to PA1 (pharyngeal arches are numbered), while this activity is lost in hoxb2a(zf)ΔPbx-Hox. GFP-expressing embryos shown in b and d are representative of the expression potential of the reporter construct in each case, as inferred from screening many (typically more than 100) injected embryos. The injection statistics for Hoxa2(m) and mutated variants in transient transgenic zebrafish embryos are provided in Supplementary Table 3, while those for the hoxb2a(zf) constructs in lamprey embryos are given in Supplementary Table 2. |
|
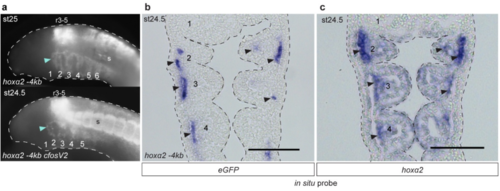
GFP reporter expression driven by lamprey hoxα2 upstream regions in transient transgenic lamprey embryos. a, Lateral views of st24-25 transient transgenic lamprey embryos showing GFP expression in rhombomeres (r), somites (s) and NC of the pharyngeal arches (numbered), driven by the hoxα2 -4kb enhancer with or without the mouse c-Fos minimal promoter. In cloning the c-Fos promoter between the lamprey enhancer and the GFP coding sequence, two alternative reporter constructs were generated: hoxα2 -4kb cFosV1 with the upstream lamprey sequence fully intact, hoxα2 -4kb cFosV2 with the 5’UTR partially removed. In both cases, the insertion of the c-Fos promoter increased levels of reporter expression but did not influence tissue-specific expression domains. GFP-expressing embryos shown are representative of the expression potential of the reporter construct in each case, as inferred from screening many (typically more than 100) injected embryos (see Supplementary Table 2 for expression statistics). b, Frontal section through a transient transgenic lamprey embryo at st24.5, revealing GFP transcripts in the NC-derived mesenchyme (arrows) of the pharyngeal arches (numbered), driven by hoxα2 -4kb. c, Frontal section through a st24.5 lamprey embryo showing endogenous hoxα2 expression. Arrows indicate elevated expression in the NC-derived mesenchyme. The pharyngeal arches are numbered. Scale bars: 100μm. Abbreviations: r, rhombomere; s, somites. |
|
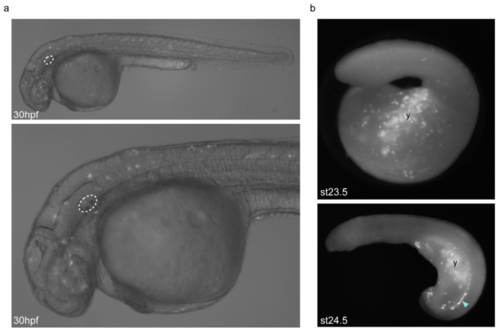
Background GFP expression driven by the empty HLC vector in zebrafish and lamprey embryos. a, Lateral views of 30hpf/prim-16 stage transient transgenic zebrafish embryos injected with the empty HLC vector using Tol2-mediated transgenesis, showing low-intensity mosaic GFP expression in multiple tissue types including neurons and muscle cells. The otic vesicle is circled. b, Lateral views of st23.5 and st24.5 transient transgenic lamprey embryos injected with the empty HLC vector using I-SceI-mediated transgenesis, showing mosaic GFP expression in the yolk (y) and ectoderm, as well as cells lying dorsal to the yolk (arrowhead). GFP-expressing embryos shown are representative of the expression potential of the reporter construct in each case, as inferred from screening many (typically more than 100) injected embryos. |