- Title
-
Multiplexed CRISPR/Cas9-mediated knockout of 19 Fanconi anemia pathway genes in zebrafish revealed their roles in growth, sexual development and fertility
- Authors
- Ramanagoudr-Bhojappa, R., Carrington, B., Ramaswami, M., Bishop, K., Robbins, G.M., Jones, M., Harper, U., Frederickson, S.C., Kimble, D.C., Sood, R., Chandrasekharappa, S.C.
- Source
- Full text @ PLoS Genet.
|
RT-PCR confirms indel mutations and identifies aberrant splice variants caused by CRISPR/Cas9 mutation. (A-D) WT and mutant allele RT-PCR products resolved on 2% agarose gel are shown in the left panels and splicing aberrations are depicted in the right panels. (A) The observed RT-PCR product for fancl_hg58 mutant was smaller in size (red arrow) than expected based on the 25 bp insertion in exon 8. The sequence of the hg58 product showed shift in its splice acceptor site resulting in loss of 48 bp sequence, that includes 23 bp from exon 8 beginning and CRISPR/Cas9 induced 25 bp insert. (B-D) Partial activation of cryptic splice site near the indel mutation was observed in fanca_hg41, fancb_hg42 and fancd1_hg45 mutant alleles. The RT-PCR products for these mutants showed two bands, one matched expected size (yellow arrows) and the other was from an altered splice product (red arrows). PCR products were cloned and sequenced to determine aberrant splice product sequence. In hg41 mutant, the indel mutation (2 bp del) in exon 12 results in partial skipping of mutated exon (B). In hg42 mutant, the indel mutation (4 bp del) in exon 1 results in partial use of new splice donor site in intron 1 (C). In hg45 mutant, the indel mutation (5 bp ins) in exon 10 results in partial use of new splice acceptor and donor in exon 10 (D). (E) The consequence on the predicted encoded protein based on the genomic indel mutation, and the observed aberrant splice product for all four mutant lines are shown. RT-PCR gel image data for all 36 alleles is shown in S3A Fig. |
|
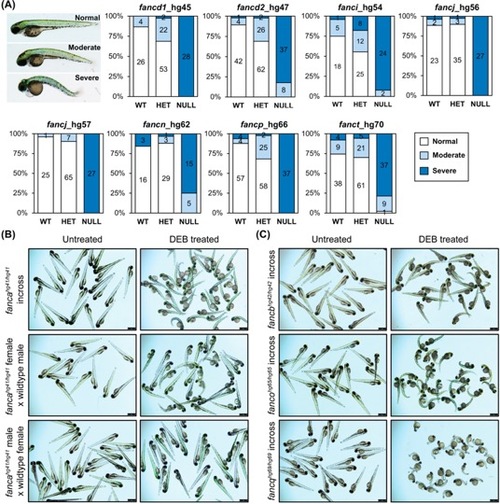
Homozygous knockout mutants are sensitive to DEB treatment. (A) Embryos obtained from inbreeding heterozygous knockouts of fancd1 (hg45; 0.9 μg/mL DEB), fancd2 (hg47; 0.9 μg/mL DEB), fanci (hg54; 0.65 μg/mL DEB), fancj (hg56 and hg57; 0.6 μg/mL DEB), fancn (hg62; 0.8 μg/mL DEB), fancp (hg66; 0.9 μg/mL DEB) and fanct (hg70; 0.8 μg/mL DEB) were treated at indicated DEB concentrations between 4–72 hpf. Treated embryos were classified based on severity of morphological changes observed into three phenotypic groups: normal (WT appearance), moderate (slight body curvature and minor edema) and severe (emaciated appearance, severe body curvature and large edema). An example image of DEB treated embryo (72 hpf) for each group is shown on the left. Distribution of each phenotypic group for a given genotype are displayed as stacked bar chart. The segments in bar show percent of embryos in each morphological group: normal (white), moderate (light blue), severe (dark blue). The number in each segment depicts the number of embryos for a given phenotypic group. (B) Maternal WT fanca transcript rescues embryos from DEB hypersensitivity. Embryos obtained from indicated fanca_hg41 breeding were treated with DEB (0.8 μg/mL). Representative images show untreated and DEB treated embryos. (C) Embryos generated from inbreeding of homozygous knockouts of fancb (hg42; 0.8 μg/mL DEB), fanco (hg65; 0.8 μg/mL DEB) and fancq (hg69; 0.5 μg/mL DEB) were treated at indicated DEB concentrations. Representative images show untreated and DEB treated embryos. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Homozygous fancp mutants are significantly smaller in body length than their siblings. Standard length measurements of fancp fish at 1 mpf (A), and 4 mpf (B). (i) Number of fish genotyped and measured for both fancp alleles: hg66 and hg67. (ii) Representative images of fancp+/+ and fancphg66/hg66 fish with red arrows marking the beginning of caudal fin used in length measurements. (iii) Data on body size measurements for fancp+/+, fancphg66/+ and fancphg66/hg66 fish. Both time points show a significant decrease in size of fancphg66/hg66 fish compared to the WT and heterozygous clutch mates (ANOVA analysis, ***p<0.001). Data for fancp_hg67 is shown in S8 Fig. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
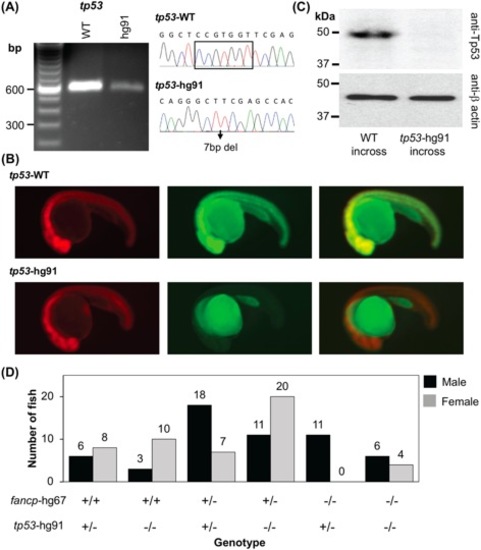
Co-mutation in tp53 gene rescues female-to-male sex reversal of fancp homozygous knockouts. (A) RT-PCR confirmed the absence of any aberrant splice variants around tp53 gene mutation. Sanger sequencing confirmed the presence of CRISPR-Cas9 introduced 7bp deletion mutation. The products were resolved on 2% agarose gel. (B) Reporter assay to check the expression of tp53 frameshift mutant. Representative images at 1 dpf of embryos co-injected with the specified reporter mRNA and TagRFP are shown as RFP (left panel), GFP (middle panel) and merged (right panel). (C) Western blot analysis shows absence of Tp53 protein in the knockout embryo extracts. Twenty-four hpf embryos collected from tp53hg91/hg91 homozygous knockout fish incross were used to check the loss of expression of Tp53 protein. Embryos obtained from TAB5 incross were used as WT controls. Expression of β-actin was used as loading control. (D) Progenies from fancphg67/+;tp53hg91/+ and fancphg67/+;tp53hg91/hg91 breeding were genotyped around 4 mpf and the sex was determined. Number of male and female fish in each genotype category is presented. The genotypes are marked on the X-axis. Among the fancp homozygous knockouts, only males were observed with fancphg67/hg67;tp53hg91/+ genotype, whereas both males and females were observed with fancphg67/hg67;tp53hg91/hg91 genotype. |
|
Homozygous knockout fish are fertile except for fancd1 and fancj knockout males. (A) Viability of embryos from outbred homozygous knockout fish at 24 hpf was determined to assess their fertility. Except fancd1 and fancj knockout males, all other knockouts (both males and females) had no fertility defect. For fancd1hg45/hg45 a small fraction of embryos (<5%) were found viable at 24 hpf. (B-C) Histology images (20x objective) of hematoxylin and eosin stained adult testis of fancd1hg45/hg45 (B) and fancjhg56/hg56 fish (C). The red arrows point to the area where mature spermatozoa reside in the testis. Empty intra-testicular ducts were observed in homozygous mutant testis (ii) but not in the heterozygous testis (i). PHENOTYPE:
|
|
RT-PCR confirms indel mutations and identifies aberrant splice variants caused by mutations. (A) RT-PCR products for all gene mutants along with WT control. The amplified products were resolved on 2% agarose gel. RT-PCR was designed to amplify the exon containing the indel mutation in knockouts and wild-type fish. Minus RT control was performed for hg50 and hg51 lines due to fancf being a single exon gene. Expected size products were observed for all mutants, except hg41 (fanca), hg42 (fancb), hg45 (fancd1), and hg58 (fancl) mutants, as denoted by the red arrows. Amplicons were sequenced to confirm the mutation, and to determine any aberrant splicing. Multiple products were sequenced after cloning into a vector. (B-E) Representative chromatograms for aberrant splice products of fancl_hg58 (B), fanca_hg41 (C), fancb_hg42 (D), and fancd1_hg45 (E). RT-PCR primers for actb2 were used as transcript control. |
|
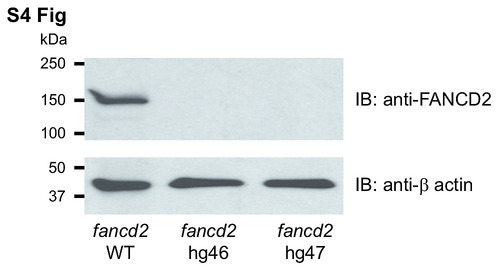
Absence of Fancd2 protein expression in fancd2 knockouts. Western blot analysis of soft tissue extracts from adult fancd2 knockout mutants using human FANCD2 antibodies. Extracts obtained from WT fish were used as controls. Expression of β-actin was used as loading control. |
|
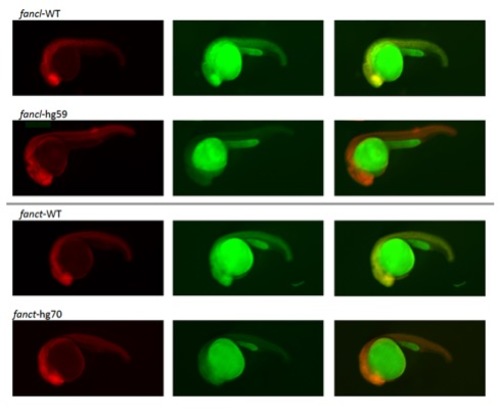
Reporter assay for expression of fance, fancf, fancg, fancl and fanct frameshift mutants. Representative images at 1 dpf of embryos co-injected with the specified reporter mRNA and TagRFP are shown as RFP (left panel), GFP (middle panel) and merged (right panel). Merged images show co-expression of the reporter (GFP) and the injection control (RFP) as yellow in the WT allele. However, in the mutant allele only the injection control (RFP) is seen as the GFP is absent due to a premature stop created by the frameshift allele. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
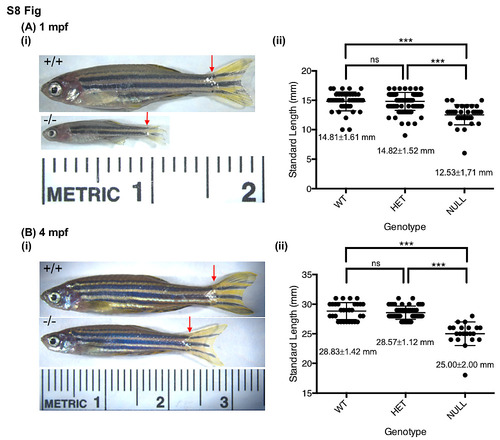
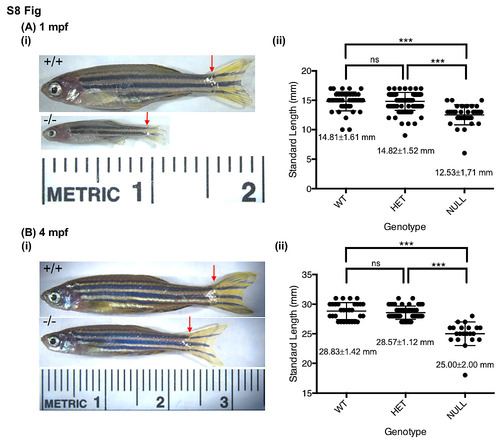
fancp-/- fish are significantly smaller in size than their siblings. Standard length measurements of fancp_hg67 fish at 1 mpf (A), and 4 mpf (B). (i) Representative images of fancp+/+ and fancphg67/hg67 fish with red arrows marking the beginning of caudal fin used in length measurements. (ii) Data on body size measurements for fancp+/+, fancphg67/+ and fancphg67/hg67 fish. Both time points show a significant decrease in size of fancphg67/hg67 fish compared to the WT and heterozygous clutch mates (Chi-square analysis, p<0.001). PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
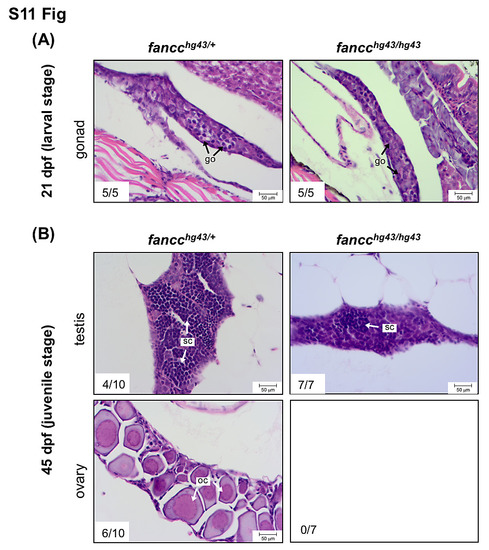
Gonadal development during larval and juvenile stages of mutant fish. Histological sections of gonads from fancc_hg43 heterozygotes and homozygotes at 21 dpf (A) and 45 dpf (B). (A) The bipotential gonads of fancchg43/+ and fancchg43/hg43 are indistinguishable at 21 dpf. (B) At 45 dpf, the gonads of fancchg43/+ exhibit continued maturation of testes or ovaries, whereas fancchg43/hg43 exhibit only testicular development. go, gonocyte; sc, spermatocyte; oc, oocyte. |