- Title
-
Grxcr1 Promotes Hair Bundle Development by Destabilizing the Physical Interaction between Harmonin and Sans Usher Syndrome Proteins
- Authors
- Blanco-Sánchez, B., Clément, A., Fierro, J., Stednitz, S., Phillips, J.B., Wegner, J., Panlilio, J.M., Peirce, J.L., Washbourne, P., Westerfield, M.
- Source
- Full text @ Cell Rep.
|
Grxcr1 Is Required for Stereocilium Widening in the Anterior Macula (A–C) Phalloidin staining of the whole anterior macula in grxcr1+/+ (A), grxcr1b1259/b1259 (B), and grxcr1b1260/b1260 mutant larvae (C). (D) Average number of hair bundles in grxcr1+/+ (n = 41 larvae), grxcr1b1259/b1259 (n = 35 larvae), and grxcr1b1260/b1260 mutant larvae (n = 32 larvae). (E) Number of TUNEL-positive cells in grxcr1+/+ (n = 18 larvae) and grxcr1−/− mutant larvae (n = 28 larvae). (F) Average hair bundle width at half their height in grxcr1+/+ (n = 40 larvae), grxcr1b1259/b1259 (n = 34 larvae), and grxcr1b1260/b1260 mutant larvae (n = 36 larvae). Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01; 5 dpf. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
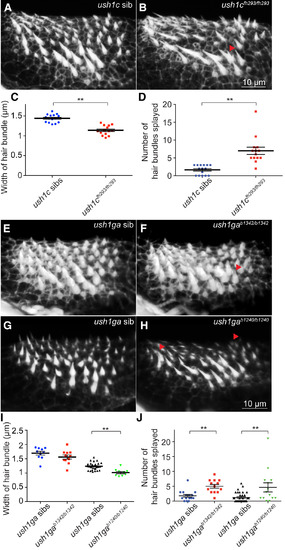
ush1c−/− and ush1ga−/− Mutants Have Dysmorphic Hair Bundles (A and B) Phalloidin staining of the anterior macula in ush1c siblings (A) and ush1cfh293/fh293 mutant larvae (B). The red arrowhead points to splayed hair bundles. (C) Average width of hair bundles at half their height in ush1cfh293 siblings (n = 15 larvae) and ush1cfh293/fh293 (n = 14 larvae). (D) Number of hair bundles splayed in ush1cfh293 siblings (n = 15 larvae) and ush1cfh293/fh293 (n = 14 larvae). (E–H) Phalloidin staining of the anterior macula at in ush1ga siblings (E and G), ush1gab1342/b1342 (F), and ush1gab1240/b1240 mutant larvae (H). (I) Average hair bundle width at half their height in ush1gab1342 siblings (n = 10 larvae), ush1gab1342/b1342 (n = 11 larvae), ush1gab1240 siblings (n = 28 larvae), and ush1gab1240/b1240 mutant larvae (n = 9 larvae). (J) Number of hair bundles splayed in ush1gab1342 siblings (n = 15 larvae), ush1gab1342/b1342 (n = 13 larvae), ush1gab1240 siblings (n = 35 larvae), and ush1gab1240/b1240 mutant larvae (n = 14 larvae). Data are represented as mean ± SEM. ∗∗p < 0.01; 5 dpf. PHENOTYPE:
|
|
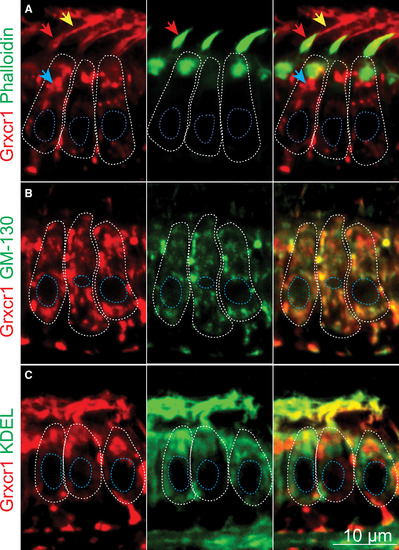
Grxcr1 Localizes in the Golgi Apparatus and ERGIC of the Hair Cells (A–C) Confocal sections of the anterior macula. Shown are partial colocalization of Grxcr1 (red) and phalloidin (A), GM130 (B), or KDEL (green, C). Individual hair cells are outlined in white. Blue, red, and yellow arrows show Grxcr1 localization in the cell body, hair bundles, and kinocilia, respectively. The confocal sections were selected based on the localization of the organelles of interest (hair bundles, Golgi, and ER), hence the differences in Grxcr1 localization. Nuclei are outlined in blue; 5 dpf. EXPRESSION / LABELING:
|
|
The Integrity of the cis-Golgi and Abundance of Ush1c and Ush1ga Are Affected in grxcr1−/− Mutants (A and B) Immunolabeling of GM130 in the anterior maculae of grxcr1+/+ (A) and grxcr1−/− (B). (C) Quantification of GM130 signal intensity in grxcr1+/+ (n = 29 larvae) and grxcr1−/− (n = 29 larvae) anterior maculae. (D and E) Immunolabeling of Ush1c in the anterior maculae of grxcr1+/+ (D) and grxcr1−/− larvae (E). (F) Quantification of Ush1c signal intensity in the anterior macula of grxcr1+/+ (n = 37 larvae) and grxcr1−/− larvae (n = 37 larvae). (G and H) Immunolabeling of Ush1ga in the anterior maculae of grxcr1+/+ (G) and grxcr1−/− larvae (H). (I) Quantification of Ush1ga signal intensity in grxcr1+/+ (n = 33 larvae) and grxcr1−/− larvae (n = 36 larvae). (J and K) Immunolabeling of Ush1g in the anterior maculae of grxcr1+/+ (J) and grxcr1−/− larvae (K). (L) Quantification of Ush1g signal intensity in grxcr1+/+ (n = 14 larvae) and grxcr1−/− larvae (n = 15 larvae). Data were normalized to grxcr1+/+ and are represented as mean ± SEM. ∗p < 0.05; 5 dpf. |
|
Grxcr1 Destabilizes Ush1c-Ush1ga Interactions (A and B) Proximity ligation assay for Ush1c and Ush1g in the anterior maculae of grxcr1+/+ (A) and grxcr1−/− larvae (B) at 5 dpf. (C) Quantification of Ush1c-Ush1g interactions in grxcr1+/+ (n = 44 larvae), grxcr1−/− larvae (n = 45 larvae), and grxcr1+/+ larvae that were incubated with only one primary antibody (control, n = 11 larvae). Data were normalized to grxcr1+/+. Individual hair cells are outlined in white. Nuclei are outlined in blue. (D) MDCK cells were transfected with ush1c-HA, ush1ga-GFP, and grxcr1-HA (+) or not (−). Extracted proteins were immunoprecipitated with an anti-GFP antibody and blotted using anti-Ush1c, anti-GFP, and anti-HA antibodies. (E) Quantification of the Ush1c level in bound fractions in the presence or absence of Grxcr1 (n = 4 experiments). Data were normalized to input and are presented as a ratio amount of normalized Ush1c in the presence of Grxcr1 over the amount of normalized Ush1c in the absence of Grxcr1. Data are represented as mean ± SEM. ∗p < 0.05. |
|
grxcr1 is expressed during early development. Related to Figure 1. (A) grxcr1 is maternally deposited and expressed during early stages of development. RT-PCR for grxcr1 and control ef1rt. hpf: hours post-fertilization. (B) grxcr1 genomic organization diagram. grxcr1 gene is composed of 4 exons. The glutaredoxin domain is indicated in orange. (C) Mosaic image of grxcr1 in situ hybridization at 5 dpf. Red arrow points to a neuromast. Red dashed lines indicate where cross sections were done. Sections through the eye (C’), the forebrain (C’’) and the anterior macula of the inner ear (C’’’). |
|
Mechanosensation is not affected in grxcr1 mutants. Related to Figure 2. (A) Percentage of larvae responding correctly to the startle response test or with a balance defect. The number of larvae analyzed was as follow: 104 grxcr1+/+, 121 grxcr1+/b1259, 43 grxcr1b1259/b1259, 112 grxcr1+/b1260, 70 grxcr1b1260/b1260. (B) Average intensity of FM1-43 signal within the hair bundles of inner ear hair cells in grxcr1+/+ (n=20 larvae) and grxcr1-/- (n=22 larvae). Incorporation of FM1-43 in the hair bundles of the hair cells of the anterior macula at 5 dpf in grxcr1+/+ (C) and grxcr1-/- (D). (E) Average intensity of FM1-43 signal within the hair bundles of neuromast hair cells in grxcr1+/+ (n=20 larvae) and grxcr1-/- (n=22 larvae). Data were normalized to grxcr1+/+. Data are represented as mean ± SEM. |
|
Grxcr1 antibody is specific, and anti-Ush1ga and commercial anti-Ush1g antibodies recognize Ush1ga but not Ush1gb. Related to Figures 5 and 6. (A) Schematic of Grxcr1 protein showing the epitope (outlined in red) recognized by the anti-Grxcr1 antibody. (B) MDCK cells were transfected with grxcr1-HA and immuno-labelled using anti-Grxcr1 and anti-HA antibodies. Grxcr1 was detected within transfected cells as shown by the co-localization between Grxcr1 (green) and HA (red). Background at the cell membrane was also observed. (C) MDCK cells were transfected with pcDNA3.1 and immuno-labelled using anti-Grxcr1 and anti-HA antibodies. Only background at the cell membrane could be detected. Immuno-labelling using anti-Grxcr1 antibody in grxcr1+/+ (D) and grxcr1-/- mutant larvae (D’) at 5 dpf. (E) Immuno-labeling using KDEL antibody in Tg[myo6b:kdel-crimson]b1319 larvae at 5 dpf. (F) Schematic of Ush1ga protein showing the epitopes recognized by the anti-Ush1ga antibody (orange line), the mouse anti-Ush1g antibody (red line), the goat anti-Ush1g antibody (blue dotted line) and the rabbit anti-Ush1g antibody (purple line). ANK domains are indicated in green and the SAM domain is shown in pink. (G) MDCK cells were transfected with ush1ga-GFP. Ush1ga was detected within transfected cells as shown by the co-localization between Ush1ga (red) and GFP (green) signals. Background was also observed in some untransfected cells. (H) MDCK cells were transfected with ush1gb-HA. Ush1ga (red) could only be detected at background level in transfected cells (green). (I) MDCK cells were transfected with ush1ga-GFP and immuno-labelled with rabbit anti-Ush1g and anti-GFP antibodies. Ush1ga was detected in transfected cells as shown by the co-localization between Ush1g (red) and GFP (green) signals. (J) MDCK cells were transfected with ush1gb-HA and immuno-labelled with rabbit anti-Ush1g and anti-GFP antibodies. Ush1gb could not be detected in transfected cells. (K) MDCK cells were transfected with ush1ga-GFP and immuno-labelled with goat and mouse anti-Ush1g antibodies and anti-GFP antibody. Ush1ga was detected in transfected cells as shown by the co-localization between Ush1g (red-goat and blue-mouse) and GFP (green) signals. (L) MDCK cells were transfected with ush1gb-HA and immuno-labelled with goat and mouse anti-Ush1g antibodies and anti-GFP antibody. Ush1gb could not be detected in transfected cells. |