- Title
-
Mga Modulates Bmpr1a Activity by Antagonizing Bs69 in Zebrafish
- Authors
- Sun, X., Chen, J., Zhang, Y., Munisha, M., Dougan, S., Sun, Y.
- Source
- Full text @ Front Cell Dev Biol
|
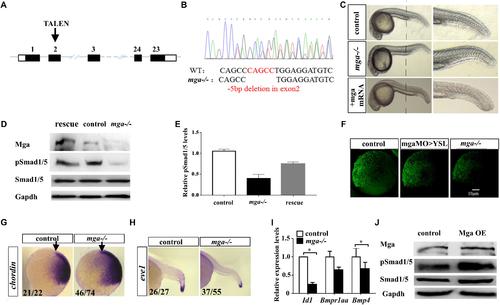
Mga mutant embryos exhibited mild dorsalized phenotype. (A) Schematic representation of the zebrafish mga gene, depicting the location of the TALEN targeting site. (B) Sequences around the TALEN targeting site, showing the TALEN-induced 5-bp deletion in mga (in red). (C) Phenotypes of 1 dpf wild type, mga mutant, and mga mutant embryos injected with 50 pg mga mRNA at one-cell-stage. The reduction of ventral tail fin was restored by injecting 50 pg mga mRNA into one-cell-stage of mga mutant embryos. Lateral view. (D) Immunoblot analysis of Mga and pSmad1/5 levels of lysates from 7 hpf control, mutant and mga mRNA restored embryos. (E) Quantification of pSmad1/5 levels of panel D based on three independent experiments. (F) pSmad1/5 gradient of wild type, mga > YSL morphants, and mga mutant embryos at 7 hpf. Dorsal to the right. (G) Chordin expression in mga mutant and control embryos at shield stage. Lateral view, and dorsal to the right. (H) eve1 expression in mga mutant and control embryos at 22 hpf. Lateral view, and dorsal to the right. (I) qRT-PCR transcript analysis of the indicated Bmp target genes in control and mga mutant embryos at 8 hpf. (J) Immunoblot analysis of pSmad1/5 levels of lysates from 8 hpf control and Mga overexpressing (OE) embryos. All experiments were performed in technical triplicate and are representative of multiple experiments. ∗p < 0.05. EXPRESSION / LABELING:
|
|
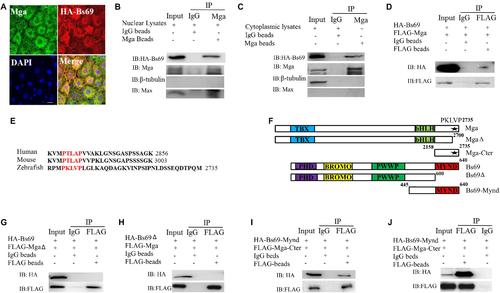
Mga interacts with Bs69. (A) Confocal images of Mga and HA-Bs69 proteins in 7 hpf embryos. Scale bar 10 μm. (B) Co-immunoprecipitation of Mga and HA-Bs69 in nuclear lysates from 8 hpf embryos. (C) Co-immunoprecipitation of Mga and HA-Bs69 in cytoplasmic lysates from 8 hpf embryos. (D) Co-immunoprecipitation of HA-Bs69 and FLAG-Mga in 293T cells. (E) Identification of a conserved PXLXP motif in human, mouse and zebrafish MGAs at the end of the C-terminal region. The PXLXP conserved motif were highlighted in red. (F) Schematic representation of the full-length and truncated Mga and Bs69 constructs as indicated. (G) FLAG-MgaΔ did not immunoprecipitate with HA-Bs69 in 293T cells. (H) FLAG-Mga did not immunoprecipitate with HA-Bs69Δ in 293T cells. (I) FLAG-Mga-Cter immunoprecipitated with HA-Bs69-Mynd in 293T cells. (J) Co-immunoprecipitation assay of the in vitro translated FLAG-Mga-Cter and HA-Bs69-Mynd. |
|
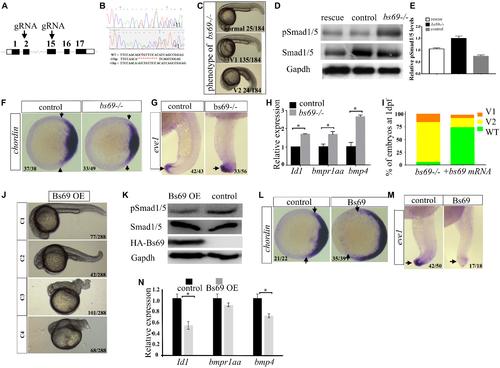
Bs69 negatively regulates Bmp signaling. (A) Schematic representation of bs69 gene, showing the CRISPR/Cas9 targeting site in exon 15. (B) Sequences of the two bs69 mutants at the CRISPR targeting site. CRISPR-induced 11 bp deletion is highlighted in red; CRISPR-induced 1 bp insertion is highlighted in green. (C) The phenotypes of bs69 mutant embryos at 1 dpf. V1: V1 ventralized phenotype; and V2: V2 ventralized phenotype, according to the DV patterning index (Kishimoto et al., 1997). (D) Immunoblot analysis of pSmad1/5 levels in lysates from 8 hpf control, bs69 mutant, and Bs69 restored embryos. (E) Quantification of pSmad1/5 levels from panel D. (F) Chordin expression in bs69 mutant and control embryos at shield stage. Animal view, and dorsal to the right. (G) eve1 expression in bs69 mutant and control embryos at 22 hpf. lateral view, and dorsal to the right. (H) qRT-PCR analysis of the Bmp target genes in 8 hpf bs69 mutant and control embryos. (I) One-cell-stage bs69 mutant embryos were injected with 50 pg bs69 mRNA, and the injected embryo at 30 hpf were scored and phenotyped according to the dorsal-ventral patterning index. (J) The phenotypes of Bs69 overexpressing embryos at 2 dpf. C1-4 dorsalized phenotypes according to DV patterning index. (K) Immunoblot analysis of pSmad1/5 levels of lysates from 8 hpf control and Bs69 overexpressing embryos. (L) Chordin expression in Bs69 overexpressing embryos at shield stage. Animal view, and dorsal to the right. (M) eve1 expression in BS69 overexpressing embryos at 22 hpf. Lateral view, and dorsal to the right. (N) qRT-PCR transcript analysis of the indicated Bmp target genes in 8 hpf control and Bs69 overexpressing embryos. All experiments were performed in technical triplicate and are representative of multiple experiments. ∗p < 0.05. EXPRESSION / LABELING:
|
|
Bs69 associates with Bmpr1a in zebrafish embryos. (A) Cellular co-localization of FLAG-Bs69 and Bmpr1a-HA in 7 hpf zebrafish embryos. Scale bar 25 μm. (B) Co-immunoprecipitation of HA-Bs69 and Bmpr1a-FLAG in cytoplasmic lysates from 7 hpf embryos. (C) Schematic representation of the full-length and truncated Bmpr1a as indicated. (D) Co-immunoprecipitation of HA-Bs69-Mynd and Bmpr1a-kinase-FLAG in 293T cells. (E) Bmpr1a-kinase-FLAG did not immunoprecipitate with HA-Bs69Δ in 293T cells. (F) Bmpr1aΔ-FLAG did not immunoprecipitate with HA-Bs69 in 293T cells. |
|
Mga binds to Bs69 and disrupts the Bs69–Bmpr1a interaction. (A) Increasing dose of pCS2-Mga (1, 2, 3 μg) along with 1 μg pCS2-HA-Bs69, 1 μg pCS2-Bmpr1a-FLAG were transiently co-transfected into 293T cells, and co-immunoprecipitation of HA-Bs69 and Bmpr1a-FLAG was analyzed. (B) Increasing dose of pCS2-Bmpr1a-FLAG (1, 2, 3 μg) along with 1 μg pCS2-HA-Bs69, 1 μg pCS2-Mga were transiently co-transfected into 293T cells, and co-immunoprecipitation of HA-Bs69 and Mga was analyzed. (C) One-cell-stage wild-type embryos injected with 50 pg mRNA encoding HA-Bs69 or mixture of mRNAs encoding both HA-Bs69 and mouse MGA, or co-injected with 50 pg mRNA encoding HA-Bs69 with 100ng pCS2-FLAG-Mga or pCS2-FLAG-MgaΔ. Immunoblot analysis of pSmad1/5 levels was performed with lysates from the injected embryos at 8 hpf. (D) Relative quantification of pSmad1/5 levels of panel C. (E) One-cell-stage wild-type embryos injected with 50 pg mRNA encoding HA-Bs69 or mixture of mRNAs encoding both HA-Bs69 and mouse or zebrafish MGAs, and the ventral tail fin phenotypes of these embryos at 48 hpf were shown. |
|
Mga localized in the cytoplasm regulates Bmp signaling. (A) Diagram showing the location of putative NLS and ENS in Mga protein. ■: NLS, ■: NES. (B) IF showing the cytoplasmic localization of FLAG-Mga-Cter in 7 hpf embryos. (C) IF showing the nuclear localization of FLAG-MgaΔNES in 7 hpf embryos. (D) FLAG-Mga-Cter but not FLAG-MgaΔNES rescued the reduced ventral tailfin phenotype of mga mutant embryos at 2 dpf. (E) pSmad1/5 levels were increased in 7 hpf mga mutant embryos injected with 100 pg FLAG-Mga-Cter but not FLAG-MgaΔNES. (F) The BRE-luc activity assay for Mga and Bs69 in C2C12 cells. (G) Cartoon model of how Mga regulates Bmp signaling. Left: Mga localized in the cytoplasm associates with Bs69 which allow Bmpr1a to phosphorylate and activate Smad1/5/8. The pSmad1/5/8 form complex with Smad4 and translocate into nuclei to regulate Bmp target gene expression. Right: In the absence of Mga, Bs69 associates with Bmpr1a and interferes with its phosphorylation and activation of Smad1/5/8 which caused reduced Bmp signaling. All experiments were performed in technical triplicate and are representative of multiple experiments. |
|
(A) The phenotypes of 1 dpf wild type embryos treated with 0.05 μM DMSO, or 0.05 μM dorsomorphin or 0.05 μM LDN193189 starting at one-cell stage, or injected with 50 pg dnBmpr1a mRNA at one-cell-stage. (B) 50 pg caBmpr1a mRNA rescued the loss of ventral tail fin phenotype of mga mutant embryos. Shown are representative embryos at 2 dpf. (C) WISH for 72 hpf embryos using mga, bs69, and bmpr1aa probes. mga and bs69 probes are DIG-labeled, whereas bmpr1aa probe is fluorescein-labeled. |
|
(A) The co-localization of Mga and HA-Bs69-Mynd in 7 hpf embryos. (B) The co-localization of zebrafish Mga and HA-Bs69 in 293T cells. (C) The co-localization of zebrafish Mga and HA-Bs69-Mynd in 293T cells. |
|
(A) Dorsalized phenotypes of HA-Bs69-Mynd overexpressing embryos at 2 dpf. C1-4 classification according to DV patterning index. (B) Western blot analysis of lysates from 8 hpf HA-Bs69-Mynd overexpressing or control embryos. (C) Tail region of 2 dpf bs69 mutant embryos injected at one-cell-stage with 50 pg mRNAs encoding HA-Bs69-Mynd or β-Gal. |
|
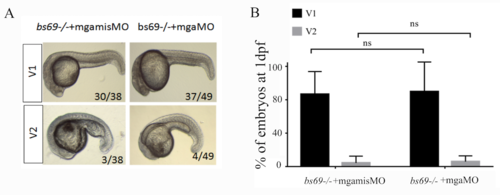
(A) DV patterning phenotypes of bs69-/- mutant embryos at 1 dpf injected with 4 ng mgamisMO or mgaMO. V1-2 classification according to DV patterning index. (B) Quantification of (A) based on three independent experiments. PHENOTYPE:
|
|
(A) The cardiac laterality defects of mga mutant or Bs69 overexpressing embryos at 1 dpf revealed by WISH using spaw, lefty2, and cmlc2 probes. (B) Percentage of embryos that exhibited cardiac laterality defects. L, left; M, middle; R, right. (C) 50 pg mouse mga mRNA partially rescued the cardiac laterality defects of mga mutants at 1 dpf; 50 pg caBmpr1a mRNA partially rescued the cardiac laterality defects of Bs69 overexpressing embryos at 1 dpf. |