- Title
-
Analysis of novel caudal hindbrain genes reveals different regulatory logic for gene expression in rhombomere 4 versus 5/6 in embryonic zebrafish
- Authors
- Ghosh, P., Maurer, J.M., Sagerström, C.G.
- Source
- Full text @ Neural Dev.
|
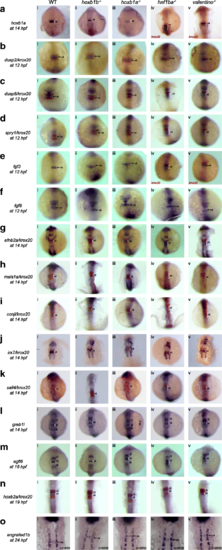
PG1 hox function is not required for expression of most r4 genes. Expression of r4 genes was assayed via ISH in (i) WT, (ii) hoxb1b mutant, (iii) hoxb1a mutant, (iv) hnf1ba mutant and (v) valentino mutant zebrafish. The genes assayed include a hoxb1a, b dusp2, c dusp6, d spry1, e fgf3, f fgf8, g efnb2a, h meis1a, i ccnjl, j irx7, k sall4, l greb1l, m egfl6, n hoxb2a and o engrailed1b. krox20 (red) which is expressed in r3 and r5, was used to assign the expression domains of several genes, as indicated. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging. Black arrows point to the r4 domain in 12hpf and 24hpf embryos. White brackets mark the normal and expanded r4 domains in embryos staged at 14hpf, 16hpf and 19 hpf EXPRESSION / LABELING:
PHENOTYPE:
|
|
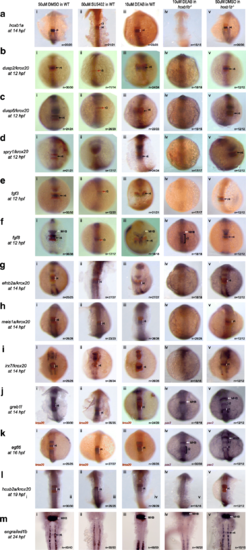
Simultaneous loss of hoxb1b and RA function disrupts expression of r4 genes. Expression of r4 genes was assayed via ISH in (i) WT embryos treated with 50uM DMSO, (ii) WT embryos treated with 50uM SU5402 (iii) WT embryos treated with 10uM DEAB, (iv) hoxb1b mutant embryos treated with 10uM DEAB and (v) hoxb1b mutant embryos treated with 10uM DMSO. The genes assayed include a hoxb1a, b dusp2, c dusp6, d spry1, e fgf3, f fgf8, g efnb2a, h meis1a, i irx7, j greb1l, k egfl6, l hoxb2a and m engrailed1b. krox20 (red), which is expressed in r3 and r5, was used to position the expression domains of several genes, as indicated. In panels (Jiv), (Jv), (Kiv) and (Kv), pax2 is the second blue marker which labels the MHB. Black arrows point to r4, red arrows to r3 and white arrows to the MHB. White and black brackets indicate r4 and r2 size, respectively. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19 hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging. This figure also shows that a subset of r4 genes is regulated by FGF signaling (Bii, Cii, Dii, Eii and Fii) and that r4 genes are not affected by the loss of RA signaling (embryos in row iii) EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression of the r5/r6 gene-set is dependent on hnf1ba and valentino. Expression of r5/r6 candidate genes was assayed via ISH in (i) WT, (ii) hoxb1b mutant, (iii) hoxb1a mutant, (iv) hnf1ba mutant and (v) valentino mutant zebrafish lines. The genes assayed include a tox3, b sema3fb, c mpz, d gas6, e celf2, f nr2f2, g col15a1b and h gbx1. krox20 (red), which is expressed in r3 and r5, was used to position the expression domains of several genes as indicated. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging. Black brackets mark the smaller r5 domain. Red bracket in (Hiii) indicates expression of gbx1 throughout the hindbrain, as a result of reappearance of gbx1 expression in r4 of hoxb1a mutants EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression of the r5/r6 gene-set requires FGF and RA signaling. Expression of r5/r6 genes was assayed via ISH in (i) WT embryos treated with 50uM DMSO, (ii) WT embryos treated with 50uM SU5402 and (iii) WT embryos treated with 10uM DEAB. The genes assayed include a tox3, b sema3fb, c mpz, d gas6, e celf2, f nr2f2, g col15a1b and h gbx1. krox20 (red), which is expressed in r3 and r5, was used to position the expression domain of some genes as indicated. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging EXPRESSION / LABELING:
PHENOTYPE:
|
|
gbx1 expression is repressed by hoxb1a in r4. a gbx1 expression is restored in hoxb1a mutant embryos. Embryos from a cross of hoxb1a heterozygous parents were assayed by ISH for gbx1 expression, producing two phenotypes (i, ii). Subsequent genotyping revealed that homozygous hoxb1a mutants express gbx1 in r4 (ii, iv). b gbx1 is initially expressed in r4 (iv), but disappears (v, vi) when hoxb1a expression is activated (i, ii, iii). c A mixture of hoxb1a and GFP mRNA was injected into 1-cell stage embryos and successfully injected embryos (identified by GFP expression) were stained for gbx1 expression at 24hpf. The observed reduction in gbx1 expression demonstrates that hoxb1a is capable of repressing gbx1 expression. d Hypothetical model depicting the potential relationship between gbx1 and hoxb1a. hoxb1a could either repress gbx1 directly (solid red T bar) or indirectly by activating a repressor (X; blue arrow) or repressing an activator (Y; orange T bar) EXPRESSION / LABELING:
PHENOTYPE:
|
|
Scheme for generating gas6 mutant line. a Schematic showing the 20 nucleotide (orange text) target site in exon 1 of gas6. CAA represents the PAM sequence (blue box) and ATG (green box) is the start codon. XcmI target sequence is indicated by the dotted red line, the red arrow denotes the cut site. b sgRNA and Cas9 mRNA was injected into 1-cell stage embryos. Injected embryos were raised to 24hpf and genomic DNA was extracted from a pool of embryos. XcmI digest of PCR products amplified from genomic DNA (extracted from injected embryos) reveal the presence of a mutation (red box in gel). c Injected embryos were raised to give rise to F0 adults. These fish were crossed with WT adults to raise the F1 generation. At 3 months age, genomic DNA was extracted from fin-clips from individual F1 fish and genotyped as in panel B. d Sequencing of F1 genomic DNA revealed transmission of four different mutant alleles (um296, um297, um298, um299), each with a different 4 nucleotide deletions (orange dashes). Each mutant allele codes for 96 out of frame amino acids (gray boxes) followed by a premature stop codon. e One quarter of the embryos collected from a cross of two heterozygous parents lack gas6 expression in r5/r6 (i). XcmI digest of PCR products amplified from genomic DNA extracted from embryos lacking gas6 expression were homozygous for mutant gas6 allele (iii, lane 5). cDNA was synthesized from total RNA extracted from WT and homozygous gas6 mutant fish. Quantitative RT-PCR using two different primer pairs (targeting the N and C termini, respectively) shows that homozygous gas6 mutants have significantly lower levels of gas6 mRNA (iv) EXPRESSION / LABELING:
PHENOTYPE:
|
|
gas6, gbx1, sall4, egfl6, celf2 and greb1l function is not required for r4-r6 formation. ISH for hindbrain markers (i) hoxb1a (blue, r4) and krox20 (red r3/5), (ii) pax2 (MHB), krox20 (r3/5) and hoxd4a (r7-anterior spinal cord), and immunostaining for neuronal markers detecting (iii) abducens motor neurons (four green dots in white boxes) in r5/r6 and (iv) Mauthner neurons (white arrows) in r4 was carried out on embryos collected from an a cross of WT fish, b cross of hoxb1a heterozygous mutants, c cross of gas6 homozygous mutants, d cross of gbx1 heterozygous mutants, e cross of greb1l homozygous mutants, f cross of a celf2 heterozygous and a homozygous mutant, g cross of egfl6 heterozygous mutants and h cross of sall4 homozygous mutants. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 14hpf and 18hpf were imaged as whole-mounts. 48hpf embryos were flat-mounted for imaging EXPRESSION / LABELING:
PHENOTYPE:
|
|
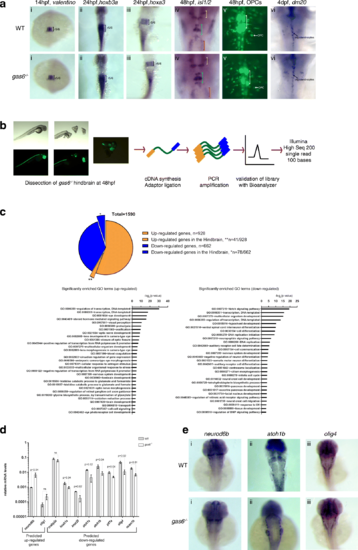
gas6 may only have subtle roles in caudal hindbrain development. a WT and gas6 mutant embryos were assayed for expression of valentino (r5/r6) (i), hoxb3a (r5-spinal cord) (ii), hoxa3 (r5/r6) (iii), islet1 (cranial nerves)(iv) and dm20 (oligodendrocyte marker) (vi) by ISH, as well as for the presence of OPCs and abducens neurons by crossing to the Tg (olig2:EGFP) vu12 line (v). In column (iv), yellow brackets mark cranial nerve V, blue brackets mark cranial nerve VII and red brackets mark cranial nerve X. White brackets indicate the presence of abducens (cranial nerve VI) in column (v). b Schemes showing RNA-seq library synthesis. Hindbrain tissue was dissected from 48 hpf gas6 mutant embryos in the olig2:eGFP background. Total RNA was collected from pools of hindbrain tissue and was used in library synthesis following the TruSeq Stranded mRNA Library Prep Kit (Illumina) protocol. c 1590 differentially expressed genes were identified from RNA-Seq where 41 out of the 928 up-regulated genes and 78 out of the 662 down-regulated genes were expressed in the hindbrain. GO terms related to Biological Processes were identified in both up-regulated and down-regulated genes using DAVID. d A subset of differentially expressed genes was validated via qPCR from independently collected hindbrain tissue samples. e ISH analysis of representative differentially expressed hindbrain genes (i) neurod6b, (ii) atoh1b and (iii) olig4 show no detectable change in expression pattern in gas6 mutant embryos EXPRESSION / LABELING:
|
|
Genotyping of embryos collected from cross of hoxb1a heterozygous parents. Several mutant lines used in this study are not viable as adults. As a result, many embryos used in assays were collected from crosses of heterozygous mutants. To ensure the presence of homozygous mutants in an assayed clutch, embryos were individually genotyped following ISH as outlined in the Methods section. Representative genotyping data for hoxb1a mutant embryos stained with (A) spry1, (B) dusp6, (C) egfl6 and (D) greb1l demonstrate that approximately one quarter of the embryos assayed are homozygous mutant (indicated with asterisks), while 100% of the clutch showed normal staining for the assayed gene. (PDF 967 kb) |
|
Expression of the r4 gene set is unaffected in hoxb1a mutants at least until 24hpf. Expression of hoxb1a (A), meis1a (B), fgf3 (C) and egfl6 (D) was assessed in wildtype (i) and hoxb1a mutant (ii) zebrafish at 24hpf. The black brackets mark r4 and dotted circles represent the otic vesicles (OV). (PDF 1051 kb) |
|
SU5402 disrupts embryogenesis in hoxb1b mutants. Wildtype (i) and hoxb1b mutant (ii) zebrafish embryos were treated with SU5402 and assayed at various developmental stages by brightfield microscopy (A, B, F, H), or ISH to detect expression of efnb2a/krox20 (C), meis1a/krox20 (D), irx7/krox20 (E) or greb1l/krox20 (G). Note that defects in development are readily detectable in hoxb1b mutants treated with 50uM SU5402 (Aii, Bii), but not in WT embryos treated with SU5402 (Ai, Bi), nor in hoxb1b mutants treated with DEAB (Biii). As a result of these severe developmental defects, hoxb1b mutant embryos treated with SU5402 showed no specific staining for the r4 genes tested. (PDF 853 kb) EXPRESSION / LABELING:
PHENOTYPE:
|