- Title
-
Coordinate regulation of retinoic acid synthesis by pbx genes and fibroblast growth factor signaling by hoxb1b is required for hindbrain patterning and development
- Authors
- Selland, L.G., Koch, S., Laraque, M., Waskiewicz, A.J.
- Source
- Full text @ Mech. Dev.
|
Hoxb1bua1006 and hoxb1asa1191 mutations. (A) The hoxb1bua1006 mutation was generated via TALEN-mediated mutagenesis and contains a 13 bp insertion 63 bp downstream of the start codon. This mutation results in a truncated 43aa protein. (B) The hoxb1asa1191 mutation contains a G to A resulting in a truncation part way through the homeodomain at amino acid 269. To determine if the protein produced by the hoxb1bua1006 allele retains any function, 200 pg of wildtype hoxb1b RNA (D) and 200 pg of mutant hoxb1b RNA containing the ua1006 mutation (E) were over expressed. In situ hybridization for the 3’UTR of hoxb1a in uninjected controls (C,F) shows normal expression in rhombomere 4. Overexpression of wildtype hoxb1b RNA results in a homeotic transformation where a duplicated region hoxb1a expression is observed in r2 (66/88 embryos affected) (D). Overexpression of hoxb1bua1006 RNA fails to cause a homeotic transformation, indicating a lack of biological activity (0/82 embryos affected) (E). Similarly, to determine if the protein produced by the hoxb1asa1191 allele retains any function, 50 pg of wildtype hoxb1a RNA (G) and 50 pg of mutant hoxb1b RNA containing the sa1191 mutation (H) were over expressed. Overexpression of wildtype hoxb1a RNA results in a homeotic transformation where a duplicated region hoxb1a expression is observed in r2 (30/34 embryos affected) (G). Overexpression of hoxb1asa1191 RNA fails to cause a homeotic transformation, indicating a lack of biological activity (0/38 embryos affected)(H). All embryos are dorsal views, anterior to the left, 18hpf. |
|
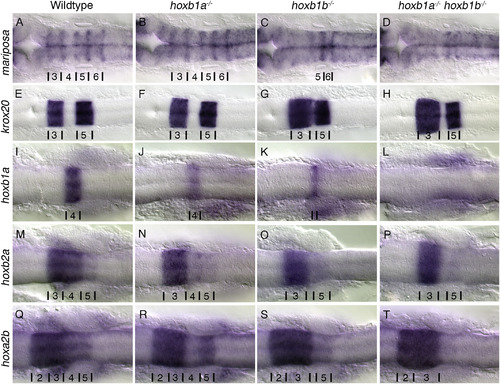
PG1 hox mutants have defects in hindbrain patterning. Rhombomere boundaries are delineated by the expression of mariposa (A-D). Wildtype (A) and hoxb1a−/− mutants (B) have normal rhombomere boundaries while hoxb1b−/− mutants (15/15 embryos)(C) have disrupted boundaries anterior to r5/6, and the boundaries in hoxb1a−/−;hoxb1b−/− mutants (3/3 embryos) (D) are not visible. Krox20 (E-H) expression in rhombomeres 3 and 5 are maintained in all embryos, however rhombomere 3 is expanded and r4 is reduced in hoxb1b−/− (29/29 embryos)(G) and hoxb1a−/−hoxb1b−/− (5/5 embryos) (H) mutants. Hoxb1a is expressed in r4 (I), hoxb2a in r3–5 (M), hoxa2b in r2-5 (Q). Expression of hoxb1a is reduced in hoxb1a−/− mutants (7/7 embryos) (J), and hoxb2a and hoxa2b expression in r4 is lost (16/16 embryos and 10/10 embryos respectively) (N,R). Hoxb1b−/− mutants have reduced expression of hoxb1a (14/14 embryos) (K), and expression of hoxb2a (7/7 embryos) (O) and hoxa2b (8/8 embryos) (S) in r4 is lost. In hoxb1a−/−;hoxb1b−/− mutants expression of hoxb1a (4/4 embryos) (L), hoxb2a (3/3 embryos) (P) and hoxa2b (3/3 embryos)(T) in r4 is lost and r5 expression of hoxb2a (3/3 embryos) (P) and hoxa2b (3/3 embryos) (T) is reduced. All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, (A-D) 22hpf, (E-T) 18hpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
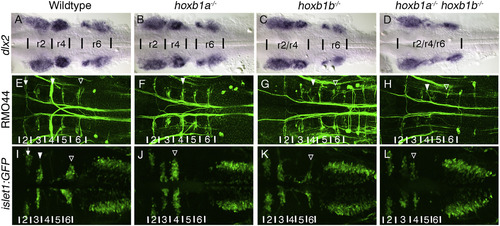
The loss of PG1 hox genes causes defects in both neural crest, and hindbrain associated neuronal populations. In hoxb1a−/− mutants three neural crest streams exit the hindbrain (B), similar to wildtype (A). The r2 and r4 streams are fused in hoxb1b−/− mutants (9/9 embryos) (C) and all three neural crest streams are less distinct in hoxb1a−/−;hoxb1b−/− mutants, with the r4 stream being reduced (8/9 embryos) (D). The Rol2 neurons (r2) (arrow), Mauthner neurons (r4) (filled arrowhead), and Mid3cm neurons (r6) (empty arrowhead) are reticulospinal neurons (labeled by RMO44) that project contralaterally across the midline (E). In hoxb1a−/− mutants the Mauthner neuron (r4) is lost (filled arrowhead), however there is a neuron resembling Rol2 in its position (10/10 embryos)(F). In hoxb1b−/− (12/12 embryos) (G) and hoxb1a−/−;hoxb1b−/− (3/3 embryos)(H) mutants, the Mauthner (filled arrowhead) and the Mid3cm (empty arrowhead) neurons are absent. Visualized with the transgenic islet1:GFP line, trigeminal BMNs differentiate to form two clusters in r2 (arrow) and r3 (filled arrowhead), the facial BMNs are born in r4 and subsequently their cell bodies migrate posteriorly to r6/7 and then laterally to their final positions (empty arrowhead) (I). The facial BMNs fail to migrate posteriorly in hoxb1a−/− mutants (11/11 embryos)(J, empty arrowhead). Hoxb1b−/− mutants specify fewer facial BMNs, however they migrate appropriately (12/12 embryos) (K, empty arrowhead). In hoxb1a−/−;hoxb1b−/− mutants there are fewer facial BMNs which fail to migrate posteriorly (5/5 embryos)(L, empty arrowhead). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior, to the left, (A–D) 22hpf, (E–L) 48hpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Hoxb1b−/−;pbx4−/− double mutants revert to the r1-like ground state. Eng2 expression in the midbrain-hindbrain boundary is observed in all embryos (A-D), however krox20 and epha4a expression are altered. As previously observed, hoxb1b−/− mutants have an expanded r3 domain and a reduced r4 domain (6/6 embryos) (B, F). In pbx4−/− mutants r3 is drastically reduced (12/12 embryos)(C,G), and in hoxb1b−/−;pbx4−/− double mutants all krox20 expression in r3 and r5 is lost (3/3 embryos)(D), while the r1 domain of epha4a expression is expanded posteriorly throughout the hindbrain (3/3 embryos)(H). All embryos have been genotyped for pbx4b557 and hoxb1bua1006. All images are dorsal views, anterior to the left, 22hpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Vhnf1 requires PG1 hox genes. Vhnf1 shows reduced expression when both hoxb1b and hoxb1a are lost (5/5 embryos)(D) compared to wildtype (A). This reduction is not observed when either hoxb1a (B) of hoxb1b (C) are lost alone. All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, 4 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
PG1 hox genes regulate vhnf1 in a retinoic acid independent mechanism. In order to determine if the decrease in vhnf1 expression observed in hoxb1a−/−hoxb1b−/− mutants is due to defects in RA, embryos were treated with 5 nM RA(A,B) or DMSO (C,D) and then vhnf1 expression was analyzed. Wildtype embryos treated with 5 nM RA (A) showed an increase in vhnf1 expression as compared to those treated with DMSO (C). In hoxb1a−/−;hoxb1b−/− mutants, there is an increase in expression of vhnf1 in embryos treated with RA (3/3 embryos)(B), as compared to controls (D), however vhnf1 expression has not recovered to wildtype levels (A). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, 4 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
RA signaling is unaffected by the loss of PG1 hox genes. The expression of aldh1a2 at 70% epiboly is unchanged in paralog group 1 mutants (B, C, D) as compared to wildtype (A). The loss of hoxb1b causes a downregulation of cyp26c1 (hoxb1b−/−: 16/17 embryos, hoxb1a−/−;hoxb1b−/−: 7/7 embryos)(G, H) and cyp26b1 (hoxb1b−/−: 20/20 embryos, hoxb1a−/−;hoxb1b−/−: 8/8 embryos)(K, L) in r4. hoxb1a−/− mutants show no reduction in the expression of cyp26c1 (F) or cyp26b1 (J) in r4. The expression of RA dependent genes, hoxd4a (M-P) and meis3 (Q-T) are unaffected by the loss of paralog group 1 hox genes. All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, (A-D) anterior to the top, 70% epiboly, (E-T) anterior to the left, 4 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
RA signaling is altered in pbx depleted embryos. To determine if the loss of hindbrain identity is due to defects in RA signaling, aldh1a2 expression was examined. Pbx4−/− mutants that are treated with pbx2/4 morpholinos have a near complete knockdown of both maternal and zygotic Pbx. At early stages of hindbrain specification there is a decrease in aldh1a2 expression in wildtype embryos treated with pbx2/4 morpholinos (C) as compared to uninjected wildtype (A) or pbx4−/− mutants (B). There is a decrease in both aldh1a2 expression level and domain in pbx4−/− mutants injected with pbx2/4 morpholinos (22/22 embryos)(D). The area of aldh1a2 expression was measured using Image J, and the average was calculated (E). Zygotic pbx4−/− mutants treated with pbx2/4 morpholinos have a 21% decrease in the area of aldh1a2 expression compared to uninjected wildtype embryos. Error bars indicate standard deviation. Uninjected wildtype: 135890.431 μm2, Uninjected pbx4−/−: 130628.408 μm2, pbx2/4MO injected wildtype: 110737.578 μm2, pbx2/4MO injected pbx4−/−: 107364.171 μm2. (ANOVA, post-hoc Tukey's: Uninjected wildtype vs pbx2/4MO injected wildtype, p-value 0.003; Uninjected wildtype vs pbx2/4MO injected pbx4−/−, p-value 0.001) All embryos have been genotyped for pbx4b557. All images are dorsal views, anterior to the top, 90% epiboly. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Hoxb1b regulates FGF signaling. Fgf3 is expressed in r4 in wildtype embryos (A) and hoxb1a−/− mutants (B). Fgf3 expression is lost in hoxb1b−/− mutants (16/16 embryos) (C) and hoxb1a−/−;hoxb1b−/− mutants (4/4 embryos) (D). Fgf8 is expressed in r4, medial r2 and the MHB (E). The loss of hoxb1a reduces the space between r2 and r4 and the MHB becomes arrowhead shaped (14/19 embryos) (F). In hoxb1b−/− mutants (19/19 embryos) (G) and hoxb1a−/−hoxb1b−/− mutants (8/8 embryos) (H) there is ectopic expression of fgf8 in r3 and reduced expression in the lateral portions of r4. The expression of FGF responsive genes, pea3 (I–L), dusp6 (M–P), spry1 (Q–T), spry2 (U–X), and spry4 (Y–B′) are unaltered from wildtype (I,M,Q,U,Y) in hoxb1a−/− mutants (J,N,R,V,Z). In both hoxb1b−/− mutants (pea3: 6/7 embryos, dusp6: 10/11 embryos, spry1: 25/25 embryos, spry2: 19/19 embryos, spry4: 23/23 embryos) (K,O,S,W,A') and hoxb1a−/−;hoxb1b−/− mutants (pea3: 6/6 embryos, dusp6: 5/5 embryos, spry1: 9/9 embryos, spry2: 5/5 embryos, spry4: 4/4 embryos) (L,P,T,X,B′) there is an increase in expression levels. Spry1 and spry2 show ectopic expression in r3 (S,T,W,X), while spry4 is ectopically expressed in r4 (A′, B′). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, 4 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
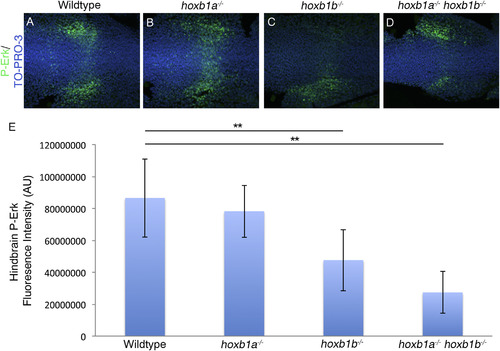
Hoxb1b regulation of FGF signaling results in decreased P-Erk localization. P-Erk is localized across the hindbrain in presumptive r4 and in lateral wings adjacent to the hindbrain in wildtype and hoxb1a−/− mutants (A-B). In hoxb1b−/− mutants (20/25 embryos)(C) and hoxb1a−/−hoxb1b−/− mutants (9/9 embryos)(D) P-Erk localization in the r4 domain across the hindbrain is reduced. Fluorescence intensity in the hindbrain was quantified using Image J, and the average fluorescence intensity was calculated (E). Hoxb1b−/− mutants have a 45% decrease in intensity while hoxb1a−/−;hoxb1b−/− double mutants have a 68% decrease in intensity. Error bars indicate standard deviation. Wildtype: 86490590AU, hoxb1a−/−: 78175326AU, hoxb1b−/−: 47583826AU, hoxb1a−/−;hoxb1b−/−: 27314030AU (ANOVA, post-hoc Tukey's: Wildtype vs hoxb1b−/−, p-value 0.007; Wildtype vs hoxb1a−/−;hoxb1b−/−, p-value 0.001) All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, 4 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Morphology of paralog group 1 hox mutants. Embryos were imaged consecutively at 22hpf (A–D), 48hpf (E-H) and 72hpf (I–L) to examine gross morphological defects. Hoxb1a−/− mutants (B, F, J) do not have any major morphological defects as compared to wildtype embryos. (A, E, I) Hoxb1b−/− mutants have a small otic vesicle at 22hpf (C) which does not persist at later stages (G, K). Hoxb1a−/− hoxb1b−/− mutants have a small otic vesicle at 22hpf (D) and develop cardiac edema at later stages (H, L). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All embryos are lateral views, anterior to the left. Hpf, hours post fertilization. PHENOTYPE:
|
|
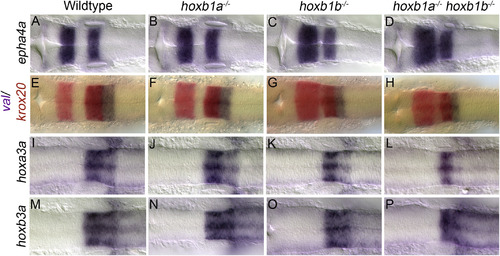
Alterations to rhombomere size in paralog group 1 hox mutants. Wildtype embryos show the normal expression patterns for epha4a in r3/5 (A), krox20 in red in r3/5 and val in purple in r5/6 (E), and hoxa3a (I) and hoxb3a (M) in r5/6. Expression of all markers are unaffected in hoxb1a−/− mutants (B,F,J,N). Hoxb1b−/− mutants have an increase in the A-P extent of r3 as observed in epha4a (C) and krox20 (G) expression. The A-P extent of r4 is reduced in hoxb1b−/− mutants as seen in the space between r3/5 staining of epha4a (C) and krox20 (G). The A-P extent of r5/6 is also slightly reduced in hoxb1b−/− mutants as seen by val (G), hoxa3a (K) and hoxb3a (O) expression. Hoxb1a−/−;hoxb1b−/− mutants have a similar expansion or r3 and reduction in the A-P extent of r4 as evidenced by epha4a (D) and krox20 (H) expression. The A-P extent of r5/6 is also reduced in hoxb1a−/−;hoxb1b−/− mutants as seen by val (H), and hoxb3a (P) expression. Hoxb1a−/−;hoxb1b−/− mutants also have a decrease in the expression of hoxa3a in r6 (L). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All embryos are dorsal views, anterior to the left, (A-D) 22hpf, (E-P) 18hpf. hpf, hours post fertilization. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Two-color analysis of rhombomere identity with cyp26c1. The expression of cyp26c1 is examined in combination with krox20. Cyp26c1 is shown in purple, while krox20 is shown in red. Krox20 expression labels r3 and r5. Cyp26c1 expression is observed in r2/4/6 in wildtype (A) and hoxb1a−/− mutants (B). The loss of hoxb1b causes a downregulation of cyp26c1 in r4 (C,D). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, 4–5 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fgf3 is reduced in hoxb1a mutants by 10 somites. Fgf3 is expressed in r4 in wildtype embryos (A) and by 10 somites, fgf3 expression is reduced in hoxb1a−/− mutants (B). All embryos have been genotyped for hoxb1asa1191. All images are dorsal views, anterior to the left, 10 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
|
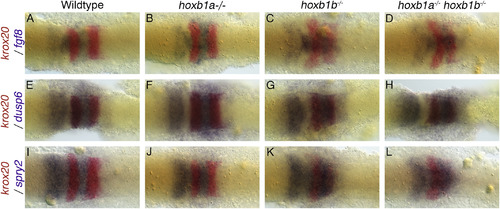
Two-color analysis of rhombomere identity with fgf8, dusp6 and spry2. The expression of representative markers is examined in combination with krox20. Fgf8, dusp6 and spry2 are shown in purple, while krox20 is shown in red. Krox20 expression labels r3 and r5. All embryos are dorsal views, anterior to the left, 4–5 somites. Fgf8 is expressed in r4, medial r2 and the MHB (A). The loss of hoxb1a reduces the space between r2 and r4 and the MHB becomes arrowhead shaped (B). In hoxb1b−/− mutants (C) and hoxb1a−/−hoxb1b−/− mutants (D) there is ectopic expression of fgf8 in r3 and reduced expression in the lateral portions of r4. The expression of FGF responsive genes, dusp6 (E-H), and spry2 (I-L), are unaltered from wildtype (E,I) in hoxb1a−/− mutants (F,J). In both hoxb1b−/− mutants (G,K) and hoxb1a−/−;hoxb1b−/− mutants (H,L) there is an increase in expression levels. Spry2 shows ectopic expression in r3 (K,L). All embryos have been genotyped for hoxb1asa1191 and hoxb1bua1006. All images are dorsal views, anterior to the left, 4 somites. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Mechanisms of Development, 150, Selland, L.G., Koch, S., Laraque, M., Waskiewicz, A.J., Coordinate regulation of retinoic acid synthesis by pbx genes and fibroblast growth factor signaling by hoxb1b is required for hindbrain patterning and development, 28-41, Copyright (2018) with permission from Elsevier. Full text @ Mech. Dev.