- Title
-
Number of inadvertent RNA targets for morpholino knockdown in Danio rerio is largely underestimated: evidence from the study of Ser/Arg-rich splicing factors
- Authors
- Joris, M., Schloesser, M., Baurain, D., Hanikenne, M., Muller, M., Motte, P.
- Source
- Full text @ Nucleic Acids Res.
|
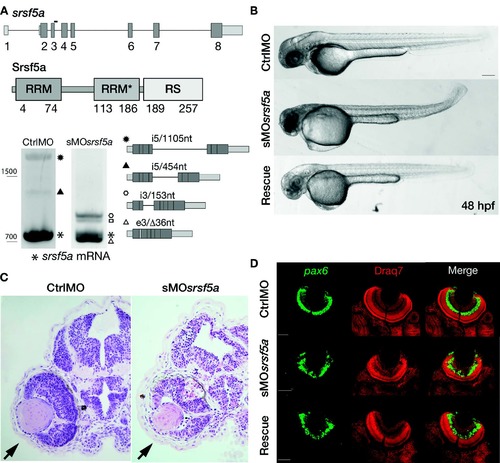
Injection of a splice site blocking MO targeting the srsf5a gene led to developmental defects. (A) srsf5a is composed of eight exons. The protein is encoded by exons 2–8 and consists of two RRM domains responsible for RNA binding and one RS domain, essential for protein–protein interactions. Three different transcripts are produced from srsf5a. Among them, two alternative transcripts retain intron 5 or a part of it (black up-pointing triangle and black dot). sMOsrsf5a was designed to target the exon3–intron3 junction. RT-PCR experiments to amplify srsf5a mRNA in control and morphant embryos at 48 hpf revealed intron3 retention (open hexagon mark), introducing a premature STOP codon into the RRM1 encoding part of the mRNA. Expression of the three normal srsf5a transcripts was strongly reduced in morphants, while MO injection also triggered the use by the splicing machinery of a cryptic splice site located in exon3, and leading to a deletion of 36 bases in the open reading frame of the srsf5a transcript (srsf5a/Δ36, open triangle mark). The resulting protein has a 12-amino acids deletion within the RRM1. The open square mark corresponds to a srsf5a transcript in which intron3 is retained and with a deletion of 36 bases in the exon3 (srsf5a/Δ36-intron3). All PCR products were identified by sequencing. (B) Zebrafish embryos injected with 3 ng of ctrlMO or sMOsrsf5a with or without a rescuing dose of srsf5a mRNA (80 pg) at 48 hpf. The defects in brain, eye and curved tail could be partially rescued by srsf5a mRNA injection. Pigmentation was not visible as embryos were treated with 1-phenyl-2-thiourea to increase their transparency. Bar: 200 μm. (C) Haematoxylin/eosin sections obtained from ctrlMO and sMOsrsf5a injected embryos revealed abnormal organization of cells in the retina and an increase of cell death in the eye and the entire brain at 48 and 72 hpf (data not shown). (D) Fluorescent in situ hybridization using a pax6b probe followed by nuclear staining using draq7® revealed the disorganization of the ganglional cell layer and of the inner nuclear layer in the retina at 72 hpf in morphants compared to control embryos. Rescue experiments allowed us to partially restore the control phenotype. Scale bar: 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
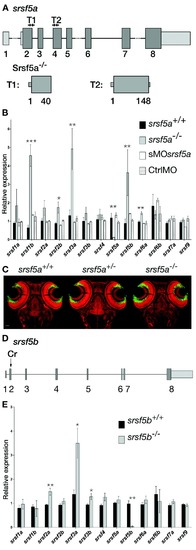
srsf5a−/−and srsf5b−/− did not show any developmental defects, but presented an overexpression of several homologous SR genes. (A) Two TALEN pairs were designed to target exon 2 or exon 4 of the srsf5a locus. TALEN pairs 1 and 2 generated of a deletion of, respectively 11 (Δ11) and 5 nt (Δ5), resulting in the production of a protein truncated in the RRM1 domain. (B) Quantitative RT-PCR to measure mRNA expression of sr genes in wild-type (wt), srsf5a mutants, morphants and ctrlMO microinjected embryos at 24 hpf. A strong decrease of srsf5a mRNA levels was observed in mutants compared to wt, suggesting the loss of Srsf5a protein in the mutant. In contrast, an upregulation of srsf1b, srsf2b, srsf3a, srsf5b and srsf6a was found. No such differences were observed in morphants compared to injected control embryos or wt. The data represent mean ± S.D. expression relative to the ef1alpha reference gene of at least three independent experiments. One-way ANOVA followed by a Tukey's multiple comparison test was used for statistical analysis. *, **, ***Mutants are statistically different from wt (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001). (C) Fluorescent in situ hybridization using a pax6b probe followed by nuclear staining using draq7® in srsf5a−/− mutants and wt. No phenotype could be detected. (D) A CRISPr (Cr) was designed to target srsf5b exon2 and allowed us to obtain three different srsf5b mutants presenting 5, 11 or 14 bases deletion. The three mutations led to the production of a truncated protein in the RRM1 domain. (E) Comparison of SR genes expression level between srsf5b homozygous mutants (including the three mutant lines) and wt embryos at 24 hpf showed an overexpression of srsf2a, srsf3a and srsf3b. A drastic decrease of srsf5b expression confirmed its depletion in mutants. All data are expressed as the mean ± SEM. A one-way ANOVA was used for statistical analysis, followed by a multiple comparison Tukey's test. *, **, *** Mutants are statistically different from wt (*P ≤ 0.05, **P ≤ 0.01). EXPRESSION / LABELING:
PHENOTYPE:
|
|
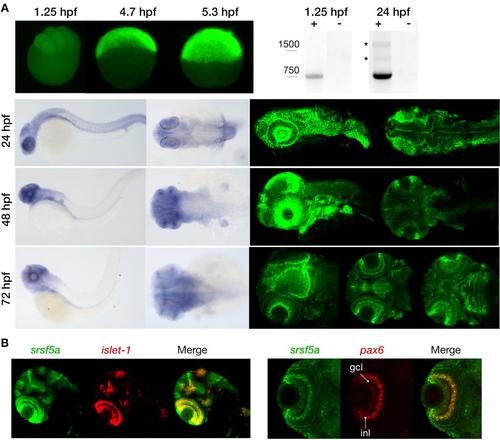
Expression pattern of the srsf5a gene. A, Visualization of maternal and zygotic srsf5a transcripts by whole mount in situ hybridization (ish) (upper left) and RT-PCR analysis (upper right). Maternal expression was detected at 1.25hpf (8 cells). A high expression of srsf5a was observed after zygotic gene activation, as shown by in situ hybridization at 4.7 (30% epiboly) and 5.3hpf (50% epiboly) (lateral views). RT-PCR analysis revealed the presence of two alternative srsf5a transcripts (*) at 24hpf that were absent at 1.25hpf. srsf5a was predominantly expressed in the brain at 24, 48 and 72hpf as determined by visible and fluorescent ish on the bottom right and left, respectively. At 24hpf, we observed an expression in the lens, the optic capsule and in the entire brain, with a more intense signal at the midbrain-hindbrain boundary. At 48hpf, transcripts for srsf5a are detected in the pectoral fins, the otic vesicles, and in the brain with a robust expression in the olfactory bulb, the iris, the optic tectum and in the cerebellum. By 72hpf, the entire brain expresses srsf5a with a more distinct expression in the developing eye, particularly in various layers of the retina, in the optic tectum and in the cerebellum. At this stage, the neuromasts and the pharyngeal region also display srsf5a expression. B, Double fluorescent ish was performed to specify the expression profile in the retina at 72hpf. srsf5a mRNA colocalizes with that of islet-1 and pax6b in the ganglional cell layer (gcl) and in the inner nuclear layer (inl) of the retina. EXPRESSION / LABELING:
|
|
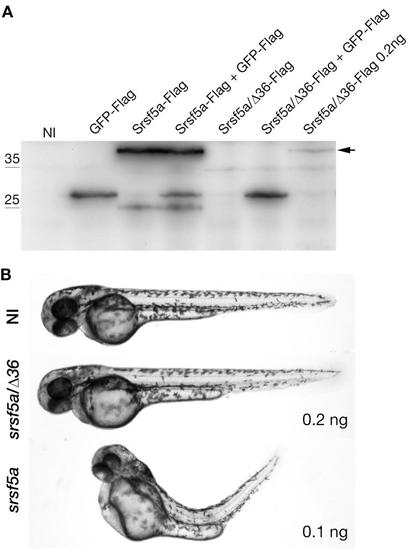
srsf5a/Δ36 cannot replace the wildtype mRNA srsf5a production. A, Expression of the Flag-tagged proteins was determined by Western blot analysis using the anti-Flag antibodies. Proteins were extracted from 24hpf uninjected embryos (NI) or injected with 50 pg of GFP-Flag, 100 pg of srsf5a-Flag, 100/200 pg of srsf5a/Δ36-Flag RNAs. Srsf5a-Flag could be visualized from 100pg of injected RNAs while Srsf5a/Δ36-Flag could be visualized when 200 pg is injected. B, Images represent uninjected embryos or injected with 100 pg of srsf5a RNAs or 200 pg of srsf5a-Δ36 RNAs at 48hpf. srsf5a injected embryos present developmental defects. |
|
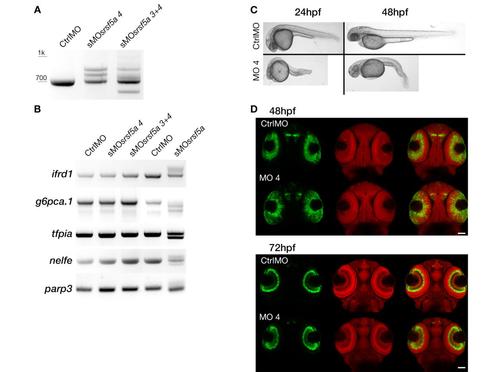
Microinjection of sMOsrsf5a4 did cause the same phenotype as sMOsrsf5a. A, RT-PCR experiments to amplify srsf5a mRNA in 48 hpf embryos injected with a control morpholino (CtrlMO), 4ng of sMOsrsf5a4 (sMOsrsf5a4) or co-injected with 3ng of sMOsrsf5a3 and sMOsrsf5a4 (sMOsrsf5a3+4) were performed. srsf5a splicing was affected in morphants compared to control as well as the srsf5a wt mRNA level. B, The sMOsrsf5a3 and sMOsrsf5a4 did not have the same effect as the sMOsrsf5a on ifrd1, g6pca.1, tfpia, nelfe and parp3 suggesting that differential splicing observed in sMOsrsf5a morphants is due to morpholino inadvertent binding and not to srsf5a knockdown. C, Injection of 4ng of sMOsrsf5a4 led to a high mortality rate (40% of death embryos at 24hpf, n>450), brain necrosis and tail defects (100% of survivable embryos at 24hpf) at 24 and 48hpf. D, Fluorescent in situ hybridization using a pax6b probe followed by nuclear staining using draq7® in 48 and 72 hpf embryos injected with the CtrlMO or the sMOsrsf5a4. No phenotype could be detected except a slight developmental delay in eye formation. |
|
Injection of sMOsrsf5a in srsf5a-/- mutant embryos led to the previously observed morphant phenotype. A, No defects could be observed in WT and srsf5a mutant embryos at 48hpf. Injection of sMOsrsf5a in wt and mutant (obtained from homozygous parents) led to developmental defects, these were more severe in mutants. B, Analysis of srsf5a transcripts produced in CtrlMO and sMOsrsf5a injected wt or srsf5a-/- mutant embryos by RT-PCR. In srsf5a mutants, the level of wt transcript is very low, while the srsf5a intron3 form is stabilized (srsf5a-i3/153nt). C, Injection of srsf5a i3/153nt at 100pg or 200pg did not lead to any developmental defect in comparison to uninjected embryos. D, Protein production from the srsf5a-i3/153nt transcript was tested by injecting a flag tagged srsf5a-i3/153nt RNA (Srsfs5-i3/153nt-Flag) at one cell stage, followed by protein extraction and western blot analysis using the appropriate antibody. No protein production could be detected. A flag tagged srsf5a RNA (Srsf5a-Flag) was used as positive control. NI = protein extract from uninjected embryos. |
|
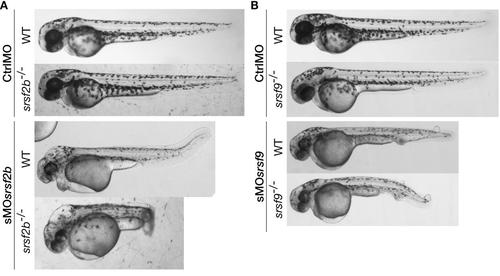
Stable loss of srsf2b or srsf9 did not lead to the previously observed morphant developmental defects. The injection of sMOsrsf2b or sMOsrsf9 in corresponding mutants recapitulated the MO-associated phenotypes. A, Injection of sMOsrsf2b in wt and srsf2b-/- led to the same developmental defects including brain oedemas. B, Knock-down of srsf9 in wt and mutants using MOs generated vascular defects and the appearance of oedemas on the body. srsf2b-/- and srsf9-/- mutants were identified among offspring from heterozygous carriers by genotyping. |
|
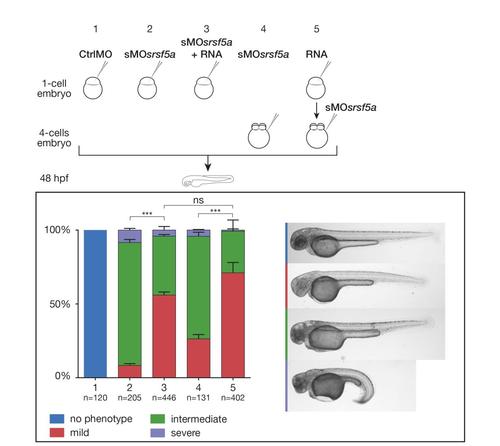
Sequential injection of RNA and sMOsrsf5a did not prevent partial rescue to occur. Based on developmental defect severity, sMOsrsf5a injected embryos were divided into three main classes; mild, intermediate and strong. Graphs representing the proportions of the three types of embryos after the indicated treatments. The total numbers of embryos pooled from n = 3 independent experiments are indicated above each bar. The graph shows mean+SEM, a Chi-square test with two degrees of freedom was performed, ***: P≤0.0001, NS: not significant. Coinjection of RNA and morpholino at one cell stage led to partial rescue. The same results were obtained when RNAs were injected at the one-cell stage and the sMOsrsf5a at the 4-cells stage in comparison to embryos injected at four cells stage with the sMOsrsf5a alone. |