- Title
-
Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time
- Authors
- Fuse, Y., Kobayashi, M.
- Source
- Full text @ Molecules
|
Regulatory mechanisms of the transcription factor-based oxidative stress response in eukaryotes. The activation mechanism of: Keap1-Nrf2/Cnc system ( |
|
Comparison of Nrf family proteins. ( |
|
A summary of the domain structure of the Nrf/Cnc transcription factors. Conservation of the Neh domains was evaluated as follows: ◎, highly conserved; ○, relatively conserved; △, partially conserved; ×, not conserved. Specific motifs were described as “highly conserved” only when the sequences were identical to mouse Nrf2. The amino acid lengths between DLG and ETGE motifs are also shown. |
|
The evolution of Nrf/Cnc transcription factors deduced from amino acid sequences. Gray bars and subscripts (s1 and s2) in Deuterostomes Nrf denote products of alternative splicing. |
|
A comparison of Keap1 proteins: ( |
|
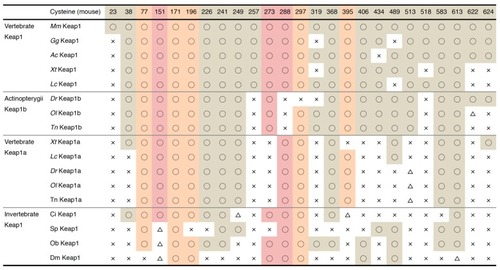
A summary of the cysteine residues of Keap1. The conservation of each cysteine is indicated as follows ○: conserved; △: not conserved but cysteine exists within three amino acids; ×: not conserved. Sensor cysteines are shaded in red, and cysteine residues conserved among Kelch family proteins in mice are shaded in orange. |
|
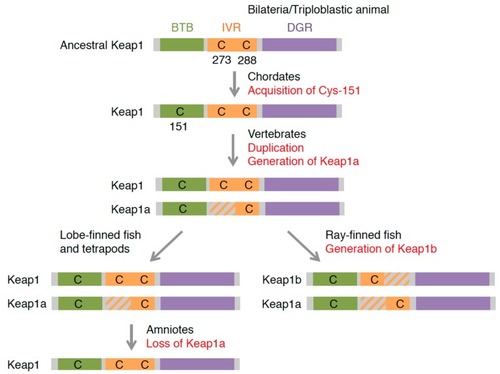
The evolution of Keap1 proteins deduced from amino acid sequences. |