- Title
-
Satellite-like cells contribute to pax7-dependent skeletal muscle repair in adult zebrafish
- Authors
- Berberoglu, M.A., Gallagher, T.L., Morrow, Z.T., Talbot, J.C., Hromowyk, K.J., Tenente, I.M., Langenau, D.M., Amacher, S.L.
- Source
- Full text @ Dev. Biol.
|
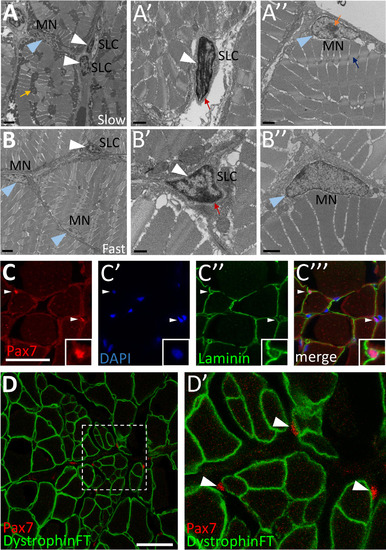
Cells with satellite cell features are present in adult zebrafish skeletal muscle. (A-B) Transmission electron microscopy (TEM) on adult zebrafish tail skeletal muscle from slow-twitch (A-A’’) and fast-twitch (B-B’’) muscle fiber domains. (A) TEM of slow muscle reveals mitochondrial-rich muscle fibers (mustard arrow points to a cluster of dark gray mitochondria), putative satellite-like cells (SLCs; white arrowheads), and myonuclei (MN; blue arrowhead points to a myonucleus). (A’) Higher magnification view of a SLC containing dense heterochromatin (white arrowhead) and surrounded by basal lamina (red arrow). (A’’) A myonucleus (MN; blue arrowhead) located within the muscle fiber membrane. Myonuclei are characterized by a lack of dense heterochromatin and often by a prominent nucleolus (orange arrow). An adjacent myofibril is indicated by a blue arrow. (B) TEM of fast muscle reveals fewer mitochondria, a putative SLC (white arrowhead), as well as myonuclei (MN, blue arrowheads). (B’) Higher magnification view of a SLC containing dense heterochromatin (white arrowhead) and surrounded by basal lamina (red arrow). (B’’) A myonucleus (MN; blue arrowhead) located within the muscle fiber membrane shows similar characteristics to slow muscle myonuclei. (C-C’’’) Pax7-positive cells (red; white arrowheads) are localized within the basal lamina of adult zebrafish skeletal muscle (anti-Laminin in green) (inset shows enlarged view of Pax7-positive cell). DAPI labeling confirms Pax7 localization in nuclei. (D-D’) Pax7-positive cells (red; arrowheads) are located outside of the muscle fiber membrane, marked by a Dystrophin FlipTrap transgenic line. (D’) Magnified view of boxed region in D. Scale bar in A, B is 2 µm, in A’, A’’, B’’ is 1 µm, in B’ is 500 nm, and in C and D is 30 µm. EXPRESSION / LABELING:
|
|
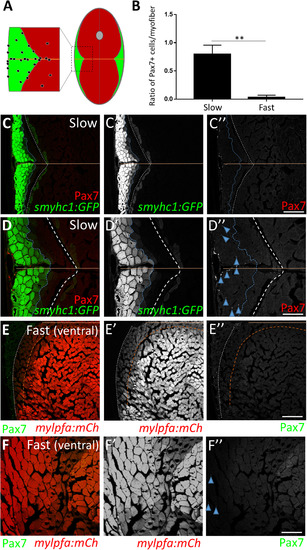
Adult zebrafish satellite-like cells are concentrated predominantly in slow muscle. (A) Schematic of cross-section through adult zebrafish trunk musculature, depicting fast-twitch (red) and slow-twitch (green) muscle fiber domains and a concentration of SLCs (black dots) within the slow muscle domain. The horizontal myoseptum is indicated by a brown line; the gray circle at the dorsal midline depicts the spinal cord. (B) The number of Pax7-positive cells, normalized to total myofiber number, is significantly higher in slow versus fast muscle; Student's t-test (p**<0.01). (C-C'') Pax7-positive cells (in red) are more common within slow muscle. Slow muscle fibers are marked with smyhc1-driven GFP (left of the dotted blue line). The boundary between slow and fast muscle fiber domains is marked by a dotted white line. The area between dotted blue and dotted white lines may represent muscle fibers with an intermediate identity. The horizontal myoseptum is indicated by a brown line. (D-D'') Higher magnification view of slow muscle. For quantification shown in B, Pax7-positive cells (D''; examples depicted by blue arrowheads) within the smyhc1:GFP-positive domain (left of the dotted blue line) were counted and normalized to the number of smyhc1:GFP-positive myofibers in the same area. (E-E'') Pax7-positive cells (in green) are rare within ventral fast muscle. Fast (ventral) muscle fibers are marked with mylpfa-driven mCherry (right of the dotted brown line). The area between dotted brown and white lines (weakly expressing mylpfa:mCherry) may represent intermediate muscle fiber identity. A dotted white line separates fast and slow muscle domains. (F-F'') Higher magnification image of fast muscle. For quantification shown in B, Pax7-positive cells (F''; blue arrowheads) within the mylpfa:mCherry-positive domain were counted and normalized to the number of mylpfa:mCherry-positive myofibers in the same area. Scale bar in C” and E” is 150 µm and in D” and F” is 75 µm. EXPRESSION / LABELING:
|
|
Satellite-like cells in adult zebrafish skeletal muscle proliferate after mechanical injury. (A) Schematic of experimental procedure, in which mechanical injury is performed by a single needle-stick into tail skeletal muscle. The approximate position of needle-stick injury in the ventral myotome is indicated in cross-sectional view. EdU was administered (gray arrowhead) by intraperitoneal injection at each post-injury time-point, allowing for EdU incorporation into S-phase cells, and fish were sacrificed 4 h later (acute EdU labeling). Tissue was collected at 2, 3, 4, 5, 6, and 7 dpi. (B) Pax7-positive cell numbers increase at the injury site and peak at 4 days post-injury (dpi). (C) Acute EdU labeling indicates robust proliferation at the injury site at 2 and 3 dpi that declines thereafter. (D) Overlap of Pax7 and EdU at 3 dpi indicates satellite-like cell proliferation post-injury. (E) Quantification of Pax7 and EdU double positive cells at each post-injury time-point reveals a peak number of proliferating satellite-like cells at 3 dpi in a 375 µm2 area encompassing the injury site; p*<0.05; p**<0.01. (F) Quantification of the percentage of Pax7-positive cells that are EdU-positive (out of total Pax7-positive cell population) at each post-injury time-point indicates that nearly 50% of Pax7-positive cells are proliferating at 2 dpi, and ~35% are proliferating at 3 dpi. The percentage of Pax7-positive cells that are EdU-positive at later post-injury time-points (4–7 dpi) is significantly less; p*<0.05; p**<0.01. One-way ANOVA followed by Tukey's multiple comparisons test was performed for statistical analyses. Scale bar in all panels is 75 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Myoblast-like cells, detected using myf5:GFP and myod:GFP transgene expression, are present post-injury in adult zebrafish skeletal muscle. (A) The myod:GFP transgenic line identifies myoblast-like cells at 4 dpi. (A'-A''') Magnified view of boxed area in A, showing myod:GFP expression (A'), EdU incorporation after an acute EdU pulse (A''), and the overlay (A''') at 4 dpi. Double-positive cells are indicated by arrowheads. (B-B''') At 4 dpi, myod:GFP-expressing cells are largely Pax7-negative (examples indicated by arrowheads). (C) Percentage of total proliferating cells (i.e. those that incorporate EdU) at 4 dpi that are Pax7-positive (25%) versus myod:GFP-positive (15%). The percentage of proliferating cells that are Pax7-positive (25%) is similar to that shown in Fig. S3I and is slightly higher than the percentage of proliferating cells that are myod:GFP-positive (15%), although the difference is not statistically significant. (D) Percentage of Pax7-expressing cells at 4 dpi that are myod:GFP-positive (10%) versus myod:GFP-negative (90%), p**<0.01. (E) myf5:GFP-positive cells are present at the injury site at 5 dpi. (E') Magnified view of boxed region in E. (E”) Magnified view of boxed region in E' reveals myf5:GFP-positive cells that are myog:H2B-mRFP-positive (arrowheads). Scale bar in A and E' is 75 µm, in E is 150 µm, and in A'-B''' and E” is 30 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
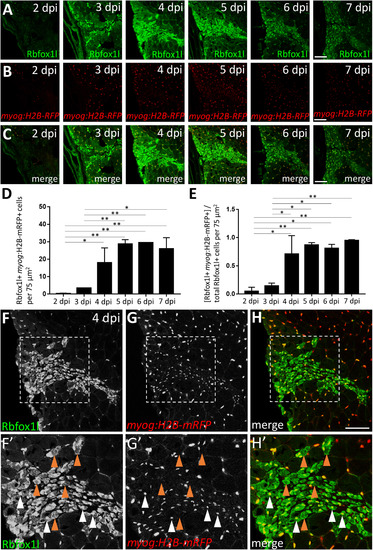
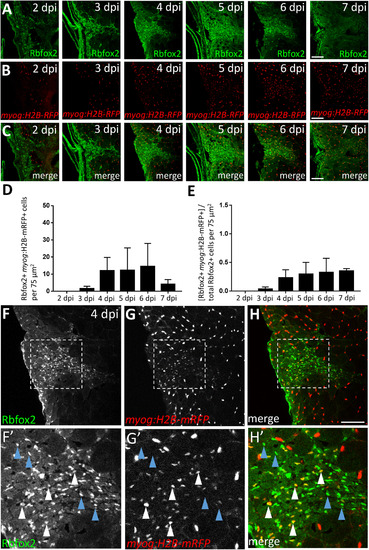
Satellite-like cells express Rbfox1l and Rbfox2 RNA-binding proteins after mechanical injury of adult zebrafish skeletal muscle. (A) Schematic of experimental procedure, which is identical to that described in Fig. 3A. (B) Rbfox1l (green) is robustly expressed beginning at 4 dpi, coinciding with the onset of new myofiber formation. Pax7 expression (overlaid in red) is the same as shown in Fig. 3C; the merge here shows overlap of Pax7 and Rbfox1l expression. The boxed region at 4 dpi is magnified in C-C''. (C-C'') Magnified view of boxed area in B, showing expression of Rbfox1l (C) and Pax7 (C'), and the overlay (C'') at 4 dpi. Examples of Pax7-positive, Rbfox1l-positive cells are indicated by purple arrowheads and of Pax7-negative, Rbfox1l-positive cells by blue arrowheads. Rbfox1l expression is observed in the cytoplasm of presumptive newly-forming myofibers (orange arrowheads indicate examples). Rbfox1l expression in uninjured areas surrounding the injury site appears less uniform in muscle fiber nuclei (compared to Fig. S6D-D'') as laser intensity was reduced to prevent over-saturation of signals within the injury site. (D) Ratio of Pax7-positive cells that are Rbfox1l-positive at post-injury time-points 2–7 dpi. (E) Rbfox2 expression (green) begins at 3 dpi and is more widespread than than Rbfox1l, closely resembling post-injury Pax7 expression (overlaid in red). The sections are alternating with those in B. The boxed region at 4 dpi is magnified in F-F''. (F-F'') Magnified view of boxed area in C, showing expression of Rbfox2 (F) and Pax7 (F'), and the overlay (F'') at 4 dpi. Examples of Pax7-positive, Rbfox2-positive cells are indicated by purple arrowheads, and of Pax7-negative, Rbfox2-positive cells by blue arrowheads. (G) Ratio of Pax7-positive cells that are Rbfox2-positive at post-injury time-points 2–7 dpi. Scale bar in B and E is 75 µm, and in C-C'' and F-F'' is 30 µm. EXPRESSION / LABELING:
|
|
Rbfox1l is expressed predominantly in the nucleus and cytoplasm of newly-forming myofibers during skeletal muscle repair. (A) Injury time-course experiment performed in the myog:H2B-mRFP transgenic line showing Rbfox1l expression at each post-injury time-point. (B) myog:H2B-mRFP expression is readily observed within the injury site by 4 dpi. (C) Overlay of Rbfox1l and myog:H2B-mRFP expression. (D) Total number of Rbfox1l, myog:H2B-mRFP double-positive cells at 2–7 dpi in a 75 µm2 area within the injury site (to exclude Rbfox1l-positive; myog:H2B-mRFP-positive myonuclei within surrounding uninjured fibers). (E) Ratio of Rbfox1l-positive cells that are myog:H2B-mRFP-positive at 2–7 dpi. One-way ANOVA followed by Tukey's multiple comparisons test was performed for statistical analyses in D and E (p*<0.05; p**<0.01). (F-H) Higher magnification view of 4 dpi injury site shown in A-C. (F'-H') Area shown is the boxed region in F-H. A majority of Rbfox1l-positive nuclei are also positive for myog:H2B-mRFP (examples indicated by white arrowheads), as expected from the extensive overlap of these two markers in differentiated muscle outside of the injury site. Rbfox1l expression in uninjured areas surrounding the injury site appears less uniform in muscle fiber nuclei in this figure (compared to Fig. S6D-D'') as laser intensity was reduced to prevent over-saturation of signals within the injury site. Some newly-forming myofibers that express Rbfox1l in the nucleus and/or cytoplasm are myog:H2B-mRFP-negative (examples indicated by brown arrowheads). Scale bar in all panels is 75 µm. EXPRESSION / LABELING:
|
|
A subset of Rbfox2-expressing cells are myog:H2B-mRFP-positive during skeletal muscle repair. (A) Injury time-course experiment performed in myog:H2B-mRFP transgenic line showing Rbfox2 expression (on alternating sections with those described in Fig. 6). (B) As shown in Fig. 6B, myog:H2B-mRFP expression is readily observed within the injury site by 4 dpi. (C) Overlay of Rbfox2 and myog:H2B-mRFP expression. (D) Total number of Rbfox2, myog:H2B-mRFP double-positive cells at 2–7 dpi. (E) Ratio of Rbfox2-positive cells that are myog:H2B-mRFP-positive at 2–7 dpi. One-way ANOVA followed by Tukey's multiple comparisons test was performed for all of the above statistical analyses; differences among time-points are not statistically significant. (F-H) Higher magnification view of 4 dpi injury site shown in A-C. (F’-H’) Area shown is the boxed region in F-H that highlights examples of Rbfox2-expressing cells that are myog:H2B-mRFP-positive (white arrowheads) and myog:H2B-mRFP-negative (blue arrowheads). Scale bar in all panels is 75 µm. EXPRESSION / LABELING:
|
|
GFP perdurance in the pax7a:GFP transgenic line implicates satellite-like cells as a source of new muscle fibers during injury-induced repair. (A) Schematic diagram depicting Pax7 protein and pax7a:GFP transgene expression in satellite-like cells and myofibers. pax7a:GFP-positive; Pax7-positive cells represent satellite-like cells. Cells that show pax7a:GFP expression within the cytoplasm but are Pax7-negative are likely differentiating or differentiated cells in which pax7a-driven GFP perdures. (B-B'') At 5 dpi, pax7a:GFP and Pax7 double-positive satellite-like cells are present at the injury site (arrowheads); however, GFP expression is also detected in Pax7-negative presumptive newly-forming myofibers. (C-E) At 4 dpi, many cells expressing MyHC (A4.1025), a differentiation marker, also express pax7a:GFP. (C'-E') Magnified view of boxed region in C-E highlights the overlap of pax7a:GFP and MyHC expression. (F-F'') pax3a:GFP and myog:H2B-mRFP transgene expression overlap in presumptive newly-forming myofibers at 4 dpi (white arrowheads), but pax3a:GFP-positive, myog:H2B-mRFP-negative are also present (mustard arrowheads). A rare presumptive newly-forming myofiber with a centrally-located nucleus is indicated by a blue arrowhead. Scale bar in B-E is 75 µm, in C', D', E' and F-F” is 30 µm. EXPRESSION / LABELING:
|
|
Pax7 function is required for skeletal muscle repair in adult zebrafish. (A) DNA lesions for pax7a alleles (oz19, oz23) and the pax7b (oz32) allele. Mutant allele sequence is shown below the wild-type sequence. Red letters and dashes indicate the lesion as well as single nucleotide changes near the CRISPR-induced frameshifting mutation. (B) CRISPR-induced frameshifting mutations in exon 2 of pax7a and pax7b cause truncations in Pax7a and Pax7b proteins, respectively, that eliminate much of the first conserved paired domain (Pax), the entire homeodomain (Hom), and the second Pax domain. The pax7aoz19 and pax7aoz23 alleles encode the first 91 of 507 amino acids of the Pax7a protein, as well as an additional 26 and 22 aberrant amino acids, respectively. The pax7boz32 allele encodes the first 81 of 510 amino acids of the Pax7b protein, as well as an additional 19 aberrant amino acids. (C-C'') pax7a+/-; pax7b+/- doubly heterozygous sibling controls show normal muscle repair at 4 dpi, indicated by expression of newly-forming myofiber markers, MyHC (A4.1025) and Rbfox1l. (D-D'') In contrast, pax7a-/-; pax7b-/- double homozygous mutants are severely deficient in newly-forming myofibers at 4 dpi, indicated by the near absence of MyHC (A4.1025) and Rbfox1l expression within the injury site. The pax7aoz23 and pax7boz32 alleles were used in these experiments. Scale bar in all panels is 75 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
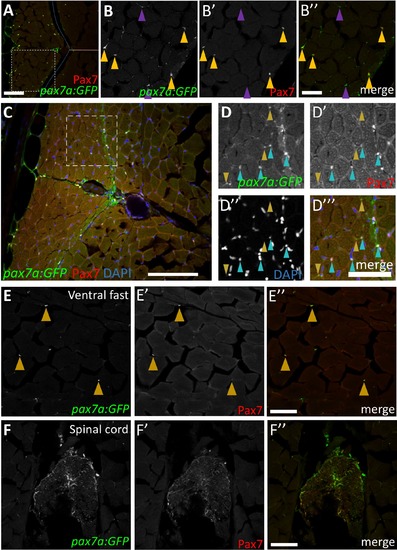
Characterization of pax7a:GFP transgene expression within the adult zebrafish myotome. (A-A'') pax7a:GFP transgenic expression overlaps with Pax7 antibody in a majority of cells within the skeletal muscle. pax7a:GFP-positive cells are also present in the skin (outer edge on left side of image). The dotted blue line represents the boundary between fast and slow muscle domains and the brown line indicates the horizontal myoseptum. pax7a:GFP is also expressed in the skin (outer edge of section). (B-B'') Higher magnification view of boxed region in A. pax7a-driven cytoplasmic GFP and anti-Pax7 labeling largely overlap (mustard arrowheads); thin cytoplasmic protrusions are also observed in cross-section. Some pax7a:GFP-positive cells are Pax7-negative (28% of total pax7a:GFP-positive population; purple arrowheads), which may represent cells with GFP-positive cytoplasm for which the nucleus is on an adjacent section. (C) pax7a:GFP-positive cells represent a subset of nuclei within the myotome. (D-D''') A majority (70%) of pax7a:GFP-positive; DAPI-positive cells are also Pax7-positive (aqua arrowheads). pax7a:GFP expression is also observed in areas lacking a corresponding nucleus (DAPI-negative; Pax7-negative) and therefore represent cytoplasmic protrusions of cells that cannot be scored for Pax7 expression (mustard arrowheads). (E-E'') pax7a:GFP-positive cells are found within the fast muscle domain, but are sparse compared to the slow muscle domain (as observed with anti-Pax7 labeling). Cells expressing both pax7a:GFP and Pax7 are indicated by mustard arrowheads. (F-F'') pax7a:GFP transgenic expression is also detected in the spinal cord. Scale bar in A”, E'', and F'' is 75 μm, in B” is 30 μm, in C is 100 μm, and in D''' is 50 μm. |
|
Characterization of pax3a:GFP transgenic expression within the adult zebrafish myotome. (A) Montage of entire cross-section showing pax3a:GFP transgene expression. In addition to satellite-like cells, pax3a:GFP is also expressed in the skin (outlining the section) and spinal cord (arrowhead). (B) Montage of entire cross-section showing pax7a:GFP transgene expression. In addition to satellite-like cells, pax7a:GFP is also expressed in the skin (outlining the section) and in a few cells within the spinal cord. (C-C''') Magnified view of ventral fast muscle region (approximate region of analysis is indicated by the box in A). Satellite-like cells expressing pax3a:GFP and Pax7 (mustard arrowheads indicate examples) are sparsely present within the ventral fast muscle region. Although the majority of pax3:GFP-expressing cells express Pax7, there are some pax3a:GFP-positive cells that are Pax7-negative (purple arrowhead) that may represent cells with GFP-positive cytoplasm for which the nucleus is on an adjacent section. Scattered EdU-positive cells are present throughout the adult myotome (white arrowhead). Scale bar in A and B is 300 μm and in C''' is 75 μm. EXPRESSION / LABELING:
|
|
By 3 dpi, a significant number of proliferating cells in the injured myotome express Pax7. (A-F) Pax7 expression (green) at post-injury time-points 2-7 dpi, with proliferating cells labeled via an acute EdU pulse (red). The image in B is also shown in Fig. 3D. (G) Pax7-positive cell number increases significantly between 2 and 4 dpi. (H) EdUpositive cell number is highest at 2 and 3 dpi and declines thereafter. (I) When the number of Pax7-positive cells is at their peak (3-4 dpi, see G), they represent ~30% of the total EdUpositive cell population, indicating that additional non-satellite-like cells are also proliferating at the injury site. One-way ANOVA followed by Tukey’s multiple comparisons test was performed for all of the above statistical analyses; p*<0.05, p**<0.01. Scale bar in F is 75 μm. |
|
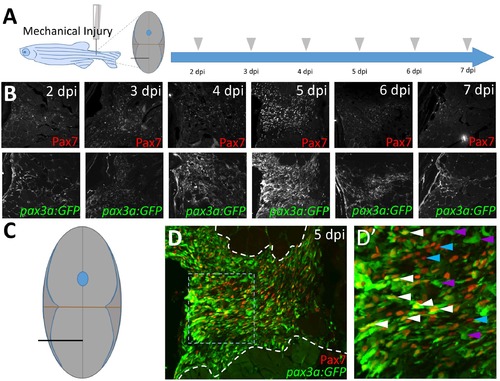
pax3a:GFP expression after injury is similar to that of Pax7, and pax3a:GFP and Pax7 are co-expressed in many cells at the peak of the satellite-like cell response. (A) Schematic of experimental procedure, in which mechanical injury is performed by a single needlestick into tail skeletal muscle. The approximate position of needle-stick injury in the ventral myotome is indicated in cross-sectional view. Tissue was collected at 2, 3, 4, 5, 6, and 7 dpi. (B) The highest number of both pax3a:GFP-positive and Pax7-positive satellite-like cells are detected at 5 dpi in this experiment. Refractive spot in the 7 dpi image is a staining artifact. (C) Schematic representation of ventral needle-stick injury site (black line). (D) Robust Pax7 and pax3a:GFP expression is detected at 5 dpi. Site of tissue repair is located between dashed white lines. (D’) Magnified view of boxed region in D. Satellite-like cells that are both pax3a:GFP and Pax7-positive (white arrowheads), pax3a:GFP-positive (purple arrowheads), and Pax7-positive (blue arrowheads) are indicated. |
|
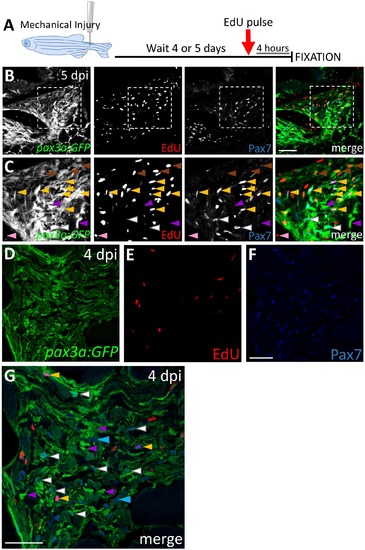
pax3a:GFP and Pax7-expressing adult zebrafish satellite-like cells proliferate after injury. (A) Schematic of acute (“pulse”) EdU labeling experimental procedure, in which mechanical injury is performed by a single needle-stick into tail skeletal muscle, followed at 4 or 5 dpi by intraperitoneal EdU injection and fixation 4 hours later. (B) Expression of Pax7 and pax3a:GFP and EdU incorporation at the injury site at 5 dpi. (C) Enlarged view from boxed region in B shows pax3a:GFP and Pax7 double positive cells at the injury site, some of which are EdU-positive (yellow arrowheads) and others EdU-negative (white arrowheads). Purple arrowheads indicate pax3a:GFP+ cells that are Pax7-negative, brown arrowheads indicate cells weakly expressing pax3a:GFP that are EdU-positive, and the pink arrowhead indicates a cell that is EdU-positive only. (D-F) pax3a:GFP, acute EdU, and Pax7 labeling at 4 dpi. (G) Overlay of channels from D-F; quantification shows that a majority (94%) of Pax7-positive cells are pax3a:GFP-positive (544/575 total cells counted). The arrowheads define cells as described above. Cells that are Pax7-positive and pax3a:GFP-negative are indicated by blue arrowheads. Scale bar in B is 75 μm and in F and G is 30 μm. |
|
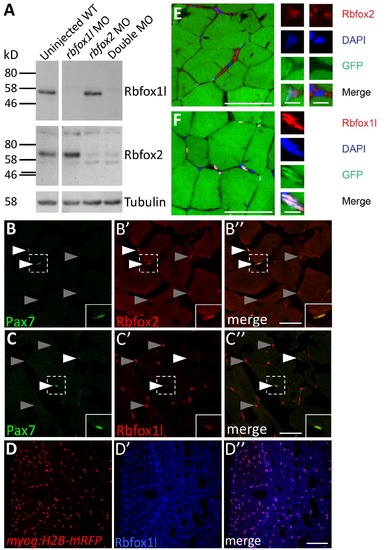
Rbfox1l and Rbfox2 are both expressed in uninjured adult zebrafish skeletal muscle, with Rbfox2 expressed mainly in satellite-like cells and Rbfox1l expressed mainly in differentiated muscle cells. (A) Immunoblot detection of Rbfox1l and Rbfox2 in 24 hours post fertilization (hpf) embryo lysates (2 embryos per lane). The Rbfox1l protein band (between 46-58 kD) is depleted after rbfox1l morpholino (MO) injection and the Rbfox2 protein band (between 58-80 kD) is depleted after rbfox2 MO injection. Both bands are depleted after co-injection of rbfox1l and rbfox2 MOs (Double MO). Tubulin expression was used as a loading control. (B-B'') Rbfox2 is expressed in Pax7-positive cells (white arrowheads; inset shows enlarged view of boxed region), and in a few other cells throughout the muscle (gray arrowheads). (C-C'') Rbfox1l is expressed in a subset of Pax7- positive cells (white arrowheads, inset of boxed region), and more prominently in nuclei that are Pax7-negative (gray arrowheads). (D-D'') Rbfox1l expression in the myog:H2B-mRFP transgenic line indicates that myonuclei are consistently Rbfox1l-positive. (E-F) Rbfox2 and Rbfox1l expression together with DAPI in the adult zebrafish myotome. Rbfox2 is localized to nuclei of some cells and is cytoplasmic in others (E). Rbfox1l is localized to nuclei based on overlap with DAPI (F). Scale bar in in B'' and C'' is 30 μm, in D'' is 75 μm, and in E, F is 50 μm in the large panels and 5 μm in the small panels. EXPRESSION / LABELING:
|
|
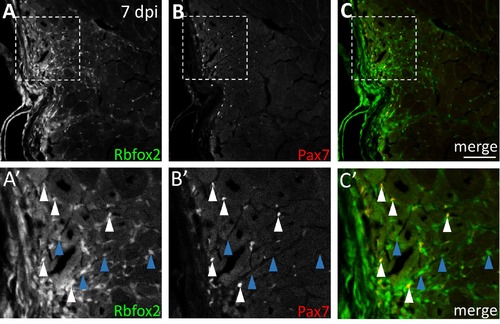
Rbfox2 shows partial overlap with Pax7 at the injury site at 7 dpi. (A-C) Some Rbfox2-positive cells at the injury site are Pax7-positive at 7 dpi. (A'-C') Higher magnification of boxed region in A-C shows Rbfox2-positive, Pax7-positive cells (white arrowheads) and Rbfox2-positive, Pax7-negative cells (blue arrowheads). Scale bar in C is 75 μm. |
|
pax7a mutants show no obvious defects in post-injury muscle repair. (A-B) Pax7 antigen is not detected in muscle or brain of pax7a-/- mutants (shown for pax7aoz19), validating loss of Pax7a in the pax7a-/- mutant, and suggesting that the anti-Pax7 antibody used may be specific for Pax7a. (C-D) Using Rbfox1l and MyHC as markers of newlyforming myofibers, no defects in post-injury repair are observed in pax7aox19-/- mutants at 5 dpi (compared to sibling controls). Scale bar in all panels is 75 μm. |
|
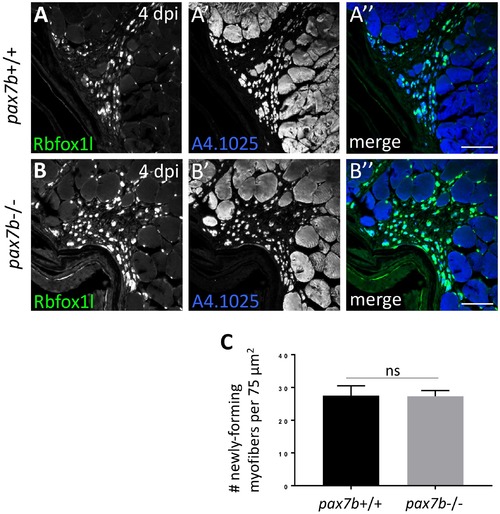
pax7b mutants show no obvious defects in post-injury muscle repair. (A-B) Using Rbfox1l and MyHC as markers of newly-forming myofibers, no defects in post-injury repair are observed in pax7boz32-/- mutants at 4 dpi (compared to sibling controls). Scale bar in all panels is 75 µm. (C) No significant difference in number of newly-forming myofibers between wild-type and pax7boz32-/- mutants at 4 dpi. Total number of myofibers counted: pax7b+/+, 165 myofibers; pax7b-/-, 164 myofibers. |
Reprinted from Developmental Biology, 424(2), Berberoglu, M.A., Gallagher, T.L., Morrow, Z.T., Talbot, J.C., Hromowyk, K.J., Tenente, I.M., Langenau, D.M., Amacher, S.L., Satellite-like cells contribute to pax7-dependent skeletal muscle repair in adult zebrafish, 162-180, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.