- Title
-
Functional antagonism of alpha-subunits of Kv channel in developing brain ventricular system
- Authors
- Shen, H., Bocksteins, E., Kondrychyn, I., Snyders, D., Korzh, V.
- Source
- Full text @ Development
|
Insertion of the GBT cassette into the second intron of zebrafish kcng4b causes a mutation. (A-D) Homozygous mutants show a clump of cells in the third ventricle (30 hpf, B,D,D′, white arrow) and hydrocephalus (48 hpf, F,G, black arrow and arrowheads). (H) TAIL-PCR mapped the insertion at position 57,917,547 on Chr. 7 in the (+) orientation (Zv9). (I) Insertion into the kcng4b locus truncates the full-length transcript. kcng4b−/− expresses the truncated transcript-kcng4b* consisting of one coding exon. (J) RT-PCR analysis demonstrates a reduction of kcng4b transcripts in kcng4b+/− and their absence in kcng4b−/− (30 hpf). Actin was detected as a loading control. (A,B) Frontal views, (C-G) lateral views, anterior to the left. E, exon; wt, wild type; A and B, primers as detailed in the Materials and Methods. |
|
kcng4b is expressed in embryos but not in adults. (A,B) RT-PCR detection of kcng4b in embryos (A,A′) but not in adults (B). (C) Antisense WISH detected expression of kcng4b in the ventricular zone (24 hpf). (C′) Zoomed image, lateral view. (C″) Zoomed image of spinal cord, dorsal view. (D) kcng4b sense probe detected no signal. (E,F) Two transverse cryosections (15 µm, oblique, midbrain). (G,H) 30-48 hpf, kcng4b transcripts in the ventricular system, otic vesicle (G″,H′), anterior and posterior eye chambers, lenses (H′), and in intersegmental vessels (H″). In G,H, eyes were removed; G′,G″ is a flat mount with yolk removed. Inset in H′ shows a zoomed image of the lens. (I) kcng4a transcripts in sensory neurons and lens at 30 hpf. (I′) Zoomed image of the head (box) with lens (arrowhead) and trigeminal ganglion (arrow). (I″) Zoomed image of the spinal cord (box), sensory Rohon-Beard neurons (RB; arrows). cc, central canal; F, forebrain; H, hindbrain; isv, intersegmental vessels; M, midbrain; ov, otic vesicle. |
|
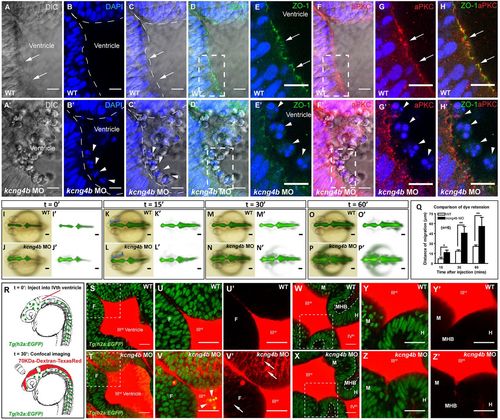
Analyses of tissue integrity. (A-H′) Confocal sections (dorsal view), in the top left corner of the anterior portion of the third ventricle. Controls (A-H) and kcng4b morphants (A′-H′). In the WT control, the apical membrane (arrow) is continuous (A); in the morphants, it is discontinuous with cells in the ventricular cavity (A′). (B′,C′) DAPI stains cell nuclei (arrowhead) in the ventricular lumen (dashed line) of kcng4b morphants. (D-H′) Apical markers ZO-1 (green, D-E′) and aPKC (red, F-G′) clearly define plasma membrane in WT, but not in kcng4b morphants. (E,E′,G,G′) Zoomed imaged of boxed areas in D,D′ and F,F′, respectively. (H,H′) Merged image from E,G and E′,G′, respectively. Images are whole mount 24 hpf embryos (dorsal view) taken with a 63× water dipping objective. (I-P′) Dye retention assay in WT and kcng4b morphants (24 hpf). Blue brackets in K,L indicate dye front. (Q) Dye leakage was quantified by measuring the extent of migration of the dye front along the blue line in K and L. Each time point represents an average of data from six independent experiments. Error bars are s.e.m. *P<0.05, **P<0.01 compared with control (unpaired Student's t-test). (R) Confocal analysis of 70 kDa Dextran leakage was performed after injection into fourth ventricle. (S-Z) Confocal section (dorsal view) of neuroepithelium 30 min after dye injection into wild-type and kcng4b MO embryo in Tg(h2b:EGFP) background at 24 hpf. Dye is seen in forebrain neuroepithelium of kcng4b MO embryos (arrows in V,V′; arrowheads show delaminated cells in V′) but not in midbrain and hindbrain neuroepithelium (X,Z). U-V′ and Y-Z′ are zoomed images of boxed areas in S,T and W,X, respectively. H, hindbrain; M, midbrain; MHB, midbrain-hindbrain boundary; ventricles are labelled with roman numerals. Scale bars: 10 µm (A-P); 25 µm (S-Z). |
|
Kcng4b regulates the brain ventricular system. (A,B) 3D reconstruction of confocal Z-scans of the brain ventricular system in vivo after injection with 70 kDa Texas Red-Dextran in dorsal view (A,A′) and tilted lateral view (B,B′). (C) Volume measurement of the brain ventricular system by 3D-reconstuction of Z-scans sections using surface function of Imaris 7.0. Data represent mean±s.e.m. of n=7; **P<0.01, unpaired t-test. (D-J) kcng4b GOF 21-24 hpf embryos. (D,E) Morphology of embryos (lateral view). (F-J) Dorsal view of embryos labelled in vivo by FITC-BODIPY-ceramide at 21-24 hpf, confocal DIC and maximal projection of Z-scans. The perpendicular line marks the widest opening of the hindbrain ventricle. (H″,I″) Confocal plane showing the ventricle with BODIPY-labelled cell membranes. Note that the rounded cells facing the lumen (arrowhead) are less abundant in kcng4b GOF (I″) compared with control (H″). (K) Number of somites at 21 and 24 hpf. Data represent mean±s.e.m.; n=25; n.s., not significant; unpaired t-test. Scale bars: 100 µm (A-B′), 50 µm (D-J). PHENOTYPE:
|
|
kcng4b modulates proliferation in neuroepithelium. (A-B′) Confocal projection view of Z-scans of embryos stained for phospho-histone 3 (PH3) and DNA (DAPI) at 24 hpf. Dorsal views, anterior to the left. (C) Proliferative index in control and kcng4b GOF embryos. Proliferative index was calculated by counting the number of PH3-positive cells and DAPI-positive neuroepithelial cells of each single confocal optical section using ImageJ cell count function, and expressed as summation of the total number of PH3-positive cells of whole Z-scans per 1000 neuroepithelial cells. ***P<0.001; unpaired t-test, n=5. (D-I) Confocal projection view of Z-scans of control and kcng4b−/− embryos stained at 24 hpf (D-E′) and 48 hpf (G-H′). (F,I) Proliferative index in controls and kcng4b mutants at 24 (F) and 48 hpf (I). ***P<0.001; unpaired t-test, n=5. Scale bars: 50 µm. |
|
kcnb1 is antagonistic to kcng4b when expressed in the brain. (A-B) Antisense Dig-RNA WISH detected kcnb1 transcripts in the ventricular system at 24 and 30 hpf. (A,B) Lateral views with anterior to the left. (A′,A″) Flat mounts of WISH-stained embryo with anterior to the left. (C,D) kcnb1 GOF 24 hpf embryos contain a clump of cells (arrow) in the third ventricle (lateral view, anterior to the left). (C′,D′) Zoomed image of the forebrain with a clump of cells (arrow). (E,F) At 48 hpf, embryos develop hydrocephalus and cardiac edema. (G) Schematic illustration of kcnb1 mutant generated using CRISPR-Cas system. Position of the target site, target sequence (blue), single guide RNA (sgRNA) sequence, PAM location (pink), isolated indel mutants (indels in red) and their respective sequences and mutant with truncated kcnb1 polypeptide are shown. In mutants #1, #2, a premature stop codon (TGA) was found. Mutant #3 is a mis-sense mutant. Mutant #1 was used for all subsequent analysis. (H,I) kcnb1 LOF results in under-inflated ventricles. (H′,I′) Ventricles filled with 70 kDa FITC-Dextran. Dorsal views with anterior to the left. cc, central canal; F, forebrain; H, hindbrain; M, midbrain; ov, otic vesicle. |
|
Schematic representation of the Tol2-mediated GBT vector composed of a protein trap cassette and a 3’ exon trap cassette. Insertion of GBT vector (detailed in upper row) into intronic sequence of a gene disrupts native splicing of the gene. The artificial splice donor (SD) and splice acceptor (SA) in the GBT vector fuse with upstream SD and downstream SA, respectively. This results in truncation of the native protein (illustrated in bottom row) Abbreviations: ITR: inverted terminal repeat; SA, splice acceptor: poly(A): polyadenylation signal with extra transcriptional terminator; β-actin: carp beta-actin enhancer and promoter, with noncoding exon and intron 1 sequences; SD, splice donor; E, enhancer; P, promoter. |
|
kcng4b splicing morpholino-mediated LOF closely recapitulated the mutant phenotype. (A) The schematics of kcng4b organization and the morpholino binding site. The predicted outcome of MO-mediated KD is this case could be very similar to that of mutation, i.e. truncation of the Kcng4 polypeptide after S1 domain. (B) the level of kcng4b expression in morphants was reduced in dose-dependent manner. (C) The morphant phenotype closely recapitulates that of the mutant, and the severity of phenotype increased in dose-dependent manner. |
|
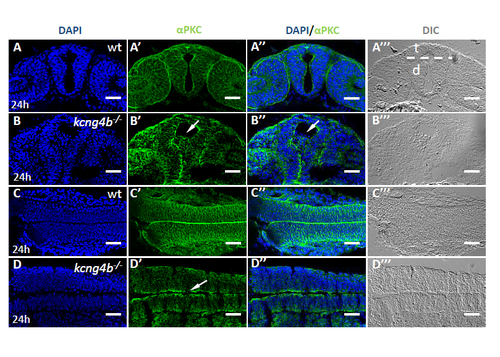
Loss of Kcng4b disrupts neuroepithelial integrity.M (A-D) Confocal image of cross-sections of embryos stained with anti-αPKC antibody (green) and DAPI (blue). A, B, forebrain (cross-section); C,D, hindbrain (tangential section). A,C , controls, B,D, kcng4b -/-. Arrow in B’, cells in the ventricle; D’, discontinuous labeling. Scale bar: 50 μm. |
|
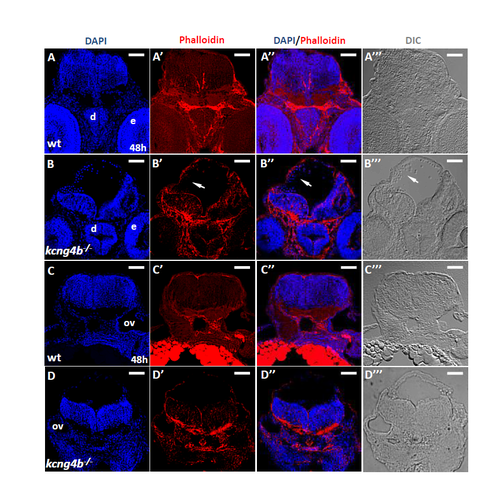
Apical/basal polarity is affected in cells that dissociated from the ventricular zone in kcng4b mutant embryos at 48 hpf. A, B midbrain (cross section near the eyes). C, D – hindbrain (cross section around otic vesicles) A, C – wild-type; B, D – kcng4b mutant embryos. Immunostaining by phalloidin (Red), counterstained with DAPI and the same section shown under DIC microscope. Scale bar: 50 μm. |
|
Overexpression of hkcng4-EGFP in zebrafish embryos blocked ventricle formation. (A, B) Injection of hkcng4-EGFP mRNA into zebrafish embryos caused absence of brain ventricles. Dorsal view of brain ventricle. Scale bar: 100 μm. (C) Comparison of number of embryos with absence of brain ventricles by injection of zebrafish kcng4b or hkcng4-EGFP mRNA. |
|
Similar expression patterns of kcng4b and kcnb1 at 72 hpf. (A-D) Composite images of two optic planes of whole-mount in situ hybridization using antisense Dig-labelled RNA probes against kcng4b and kcnb1. A, B, dorsal planes; C, D, ventral planes. E, F, lateral view after removal of eyes. Abbreviation: c, cerebellum; ht, hypothalamus; l, lens; ot, optic tectum, IIId, dorsal III-rd ventricle; IIIv, ventral III-rd ventricle; IV, IV-th ventricle. |
|
kcnb1 mutant embryos also displayed epiboly phenotype at early developmental stages. (A) The epiboly phenotype in the kcnb1 mutants. (B) The summary of phenotype scoring in kcnb1-/- mutants. PHENOTYPE:
|