- Title
-
Glypican4 Modulates Lateral Line Collective Cell Migration Non Cell-Autonomously
- Authors
- Venero Galanternik M., Lush, M.E., Piotrowski, T.
- Source
- Full text @ Dev. Biol.
|
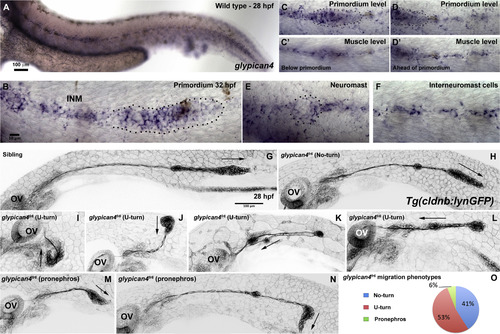
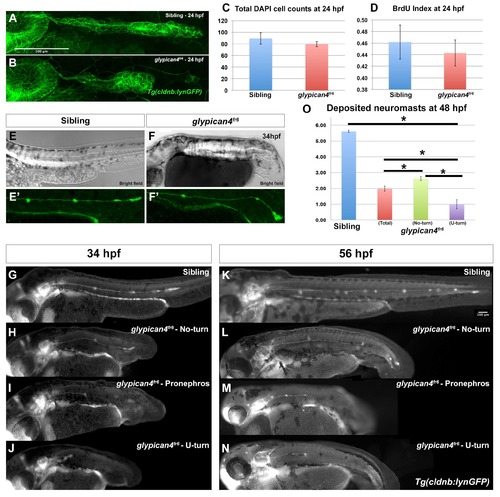
Mutations in glypican4 cause primordium migration defects. Anterior is to the left in all figures. (A) Lateral view of a 28 hpf wild type zebrafish embryo stained for glypican4 mRNA. (B) glypican4 in situ hybridization in a 32 hpf migrating primordium showing expression in the trailing region of the primordium and in the interneuromast cells (INM) deposited behind it. (C-D′) glypican4 is also expressed in muscle underlying the primordium all the way to the tail tip. (E) glypican4 is expressed in a deposited neuromast and (F) interneuromast cells at 32 hpf. (G-N) Embryos in the Tg(cldnb:lynGFP) background. The otic vesicle (OV) is used as a reference for primordium position along the embryo trunk. (G) Lateral view of a wild type 28 hpf embryo showing proper migration of the lateral line primordium. (H) Lateral view of a 28 hpf glypican4fr6‘No-turning’ mutant. (I-L) About 40% of glypican4fr6 mutant primordia possess a severe migration defect where the primordium makes a U-turn and migrates back toward the otic vesicle. This behavior occurs either very close to the otic vesicle without much migration toward the tail (I-J) or later when the primordium has already migrated partially along the tail (K-L). (M-N) Another less common phenotype observed in glypican4fr6 mutants are primordia that shift their migratory path toward the pronephros. This phenotype can occur soon after migration has started (M) or when the primordium has already migrated a considerable distance (N). (O) Quantification of the observed glypican4fr6 mutant phenotypes is represented in (H-N). |
|
Wnt/β-catenin target genes are not affected but some Fgf target genes are slightly reduced in glypican4fr6 mutants. Expression of the Wnt/β-catenin target genes lef1 (A-B), fgf10a (C-D), fgf3 (E-F), sef (G-H), and the Fgf target pea3 (I-J) and dkk1b (K-L) in 32 hpf sibling embryos and glypican4fr6 mutants respectively. pea3 is reduced, whereas dkk1b is normal. |
|
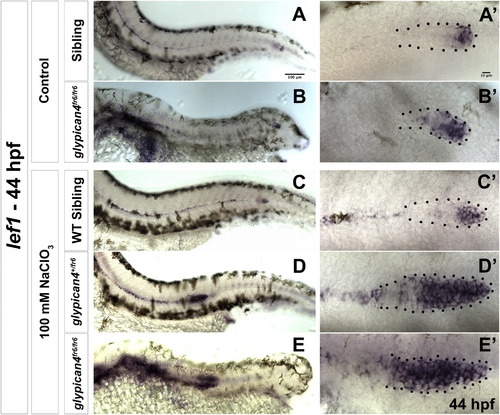
glypican4 acts redundantly with other HSPGs in the primordium. (A-B′) Lateral views of sibling (A) and glypican4fr6 mutant (B) embryos stained for the Wnt/β-catenin target lef1. (A-B) Sibling and glypican4fr6 mutant embryos show normal expression of lef1 restricted to the leading region of the primordium. (A′-B′) show magnifications of the primordia in (A-B). (C-E′) Lateral views of a sibling (C), a glypican4+/fr6 heterozygous (D) and a homozygous glypican4fr6 mutant (E) treated with a suboptimal dose of Sodium Chlorate (100 mM) and stained for lef1. The treatment does not affect expression of lef1 in wild type primordia (C′) but lef1 expression expands in both heterozygous glypican4+/fr6 and homozygous glypican4fr6 mutants (D′ and E′). |
|
Dkk1b is able to inhibit Wnt/β-catenin and Fgf signaling in the absence of glypican4. (A-D) Expression of the Wnt/β-catenin target lef1 in non-transgenic siblings (A) and glypican4fr6 mutants (B) in the leading region of the primordium. (C-D) After heat shock, 28–30 hpf transgenic siblings (C) and glypican4fr6 mutants (D) lose expression of lef1 indicating that Dkk1b is functional and inhibits Wnt target genes. (E-H) The Fgf target pea3 is expressed in the trailing region of the primordium in non-transgenic control siblings (E) and glypican4fr6 mutants (F) and is downregulated after induction of Dkk1-GFP in both backgrounds indicating that the inhibition of Wnt targets results in the loss of Fgf signaling (G-H). |
|
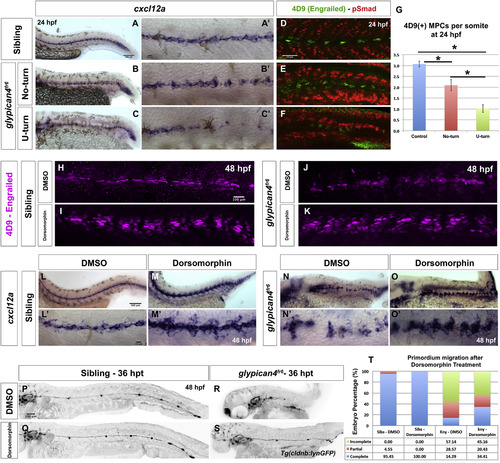
Bmp inhibition rescues Engrailed(+) muscle cells and cxcl12a expression in glypican4fr6. (A-C′) In situ hybridization shows expression of the Chemokine ligand cxcl12a in siblings (A-A′), glypican4fr6 No-turning mutants (B-B′) and glypican4fr6 U-turning mutants (C-C′) along the embryo trunk. (A′-C′) Higher magnification images of (A-C). (d-F) Engrailed (4D9) immunostaining reveals that, compared to siblings (D), glypican4fr6 No-turning mutants (E) and glypican4fr6 U-turning mutants (F) have a considerably diminished number of MPCs and MFFs. (G) Quantification of 4D9-positive muscle pioneer cells per somite at 24 hpf (p-values (t-test) for Control vs. No-turn=0.0036, Control vs. U-turn=5.09721×10−12, No-turn vs. U-turn=0.00065). Error bars represent standard error. (H-K) In control (H-I) and glypican4fr6 mutant (J-K) embryos inhibition of Bmp signaling with 10 μM Dorsomorphin induces the formation of MPCs and MFFs along the myoseptum. (L-O′) Bmp inhibition results in the upregulation of cxcl12a expression along the myoseptum in siblings (L-M′) and restores cxcl12a expression in glypican4fr6 mutants (N-O′) along the embryo trunk. (L′-O′) Higher magnification images of (L-O). (P-T) Inhibition of Bmp signaling does not affect primordium migration (P-Q) but significantly rescues the migration of glypican4fr6 mutants (R-S). (T) Quantification of primordia that have completed migration after Bmp inhibition, Fisher Exact Probability Test, Sibling-DMSO vs. Siblings-Dorsomorphin =0.31; glypican4fr6-DMSO vs. glypican4fr6-Dorsomorphin=0.04. Hours post treatment (hpt). |
|
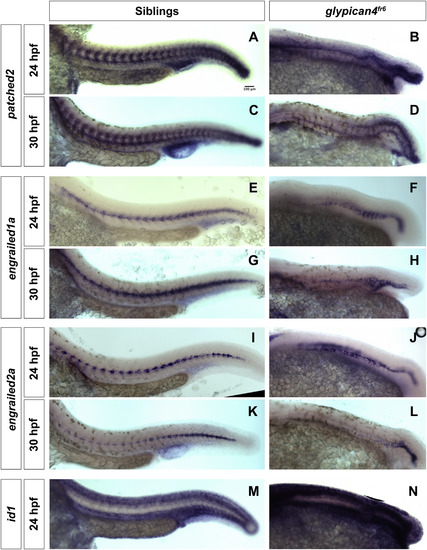
glypican4fr6 mutants show reduced expression levels of patched2, engrailed1a and engrailed2a. (A–D) 10× lateral views of the expression of the Hh target patched2 in sibling and glypican4fr6 mutant embryos at 24 and 30 hpf. (E-H) 10× lateral views of the expression of the Hh target engrailed1a in sibling and glypican4fr6 mutant embryos at 24 and 30 hpf. (I-L) 10x lateral views of the expression of the Hh target engrailed2a in sibling and glypican4fr6 mutant embryos at 24 and 30 hpf. These Hh targets show a drastic, increasing downregulation in glypican4fr6 mutants over time compared to their siblings. (M-N) 10× lateral views of the expression of the Bmp target id1 in sibling and glypican4fr6 mutant embryos at 24 hpf. This Bmp target shows a drastic upregulation in glypican4fr6 mutants compared to their siblings. |
|
glypican4fr6 mutant primordia display non-cell autonomous migration defects. (A, A′, A'') 26 hpf glypican4fr6;Tg(cldnb:lynGFP) mutant embryo into which wild type fluorescent Alexa-568 dye labeled cells were transplanted. Wild type cells are in magenta and glypican4fr6 cells are labeled green by cldnb:lynGFP. (A-A′) Wild type clones are found in the brain, skin, yolk, otic vesicle and primordium cells. (B) 40X magnification of the primordium clone boxed in (A) at 26 hpf showing magenta wild type cells in the otic vesicle (OV), lateral line ganglion and the primordium. Wild type primordium cells are not able to rescue the glypican4fr6 U-turning mutant phenotype and the primordium turns toward the ear. (C) Same primordium at 32 hpf. (D) By 48 hpf the mosaic primordium has stalled completely, irrespective of the number of wild type cells it contains demonstrating that glypican4 acts non cell-autonomously in the primordium. See Movie S3. (E-F”) glypican4fr6;Tg(cldnb:lynGFP) mutant cells (GFP+) labeled with fluorescent Alexa-568 dye were transplanted into a wild type embryo and primordium clones were screened at 26 hpf. (E-F) Shows a 10X image of the same embryo at 26 hpf and 48 hpf, respectively, in which the mutant primordium was able to complete migration and deposit neuromasts along its way to the tail tip. (B′, C′, D′, E′, F′) GFP+glypican4fr6 cells and (B”, C”, D”, E”, F”) red Alexa-568-labeled transplanted wild type cells. |
|
glypican4fr6 primordia have morphology defects and deposit fewer neuromasts. (A-B) Compared to control siblings where the primordium has an elongated ovoid shape (A), glypican4fr6 mutant primordia are more rounded and of smaller size (B). (C) Cell counts by DAPI staining show no significant differences between sibling and glypican4fr6 mutant primordia at 24 hpf (p-value (t-test)=0.327). (D) The reduced size of the 24 hpf glypican4fr6 mutant primordia is not due to reduced proliferation (p-value (t-test)=0.682). (E-F) Bright field images of a sibling control and a glypican4fr6 mutant embryo at 34 hpf depicting the embryo trunk phenotypes. (E′-F′) Same embryos represented in (E) and (F) under fluorescent light showing the lateral line primordium migrating along the trunk. No-turning glypican4fr6 mutant primordia reach the tail tip shortly after 34 hpf, whereas the sibling primordium has just passed the yolk extension. (G-J) Lateral views of 34 hpf embryos depicting representative phenotypes of sibling (G) and the three phenotypes of glypican4fr6 mutant primordia migration (H-J). (K-N) Lateral views of 56 hpf embryos depicting representative phenotypes of sibling (K) and the three phenotypes of glypican4fr6 mutant primordia migration (l-N). (O) Quantification of deposited neuromasts at 58 hpf in sibling versus glypican4fr6 mutants (siblings=5.61±0.08 vs. glypican4fr6(total)=1.98±0.17 neuromasts, n=56 embryos. sibling vs. glypican4fr6 (total) p-value=1.7×10−36; sibling vs. glypican4fr6 (No-turn) p-value=2.38×10−35; sibling vs. glypican4fr6 (U-turn) p-value=1.77×10−33). Error bars in (C), (D) and (O) represent standard errors and (*) represent statistically significantly phenotypes. PHENOTYPE:
|
|
Wnt//β-catenin signaling does not expand in glypican4fr6 mutants. (A-B) Expression of the Wnt//β-catenin ligand wnt10a is sibling (A) and glypican4fr6 mutants (B) is restricted to the leading region at 30 hpf. (C-E) The Wnt//β-catenin domain in siblings (n=5) (C) and glypican4fr6 mutants (n=7, D) is proportional to the primordium size at 28 hpf. (E) Quantification of the Wnt//β-catenin domain ratio in the primordium, p-value (t-test)=0.83. Error bars represent standard error. |
|
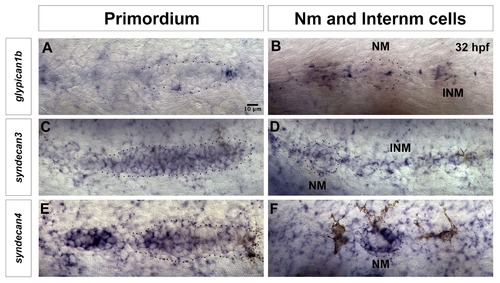
Expression of other HSPGs in lateral line cells at 32 hpf. (A) The Wnt/β-catenin target gene glypican1b is expressed in the leading region of the lateral line primordium, as well as in (B) interneuromast cells. (C–F) syndecans, are also expressed in the lateral line. (C) syndecan3 is expressed in most cells of the primordium, (except the very leading cells) and in neuromasts and interneuromast cells (D). (E) syndecan4 expression is restricted to the Fgf domain in the trailing part of the primordium and it is (F) strongly expressed in a circular pattern in the support cells of the deposited neuromast. NM: Neuromast, INM: Interneuromast cells. |
|
glypican4fr6 mutants do not show changes in primordium-expressed Chemokine receptors. (A-C) Expression of the chemokine receptor cxcr4b is not altered in 28 hpf siblings (A), glypican4fr6 No-turning (B) or glypican4fr6 U-turning mutant primordia (C). (d-F) Expression of the chemokine receptor cxcr7b is not altered in 28 hpf siblings (D), glypican4fr6 No-turning (E) and glypican4fr6 U-turning mutant primordia (F). (G) Zebrafish medusa/cxcl12at30516 mutants show primordium stalling and U-turning behavior similar to glypican4fr6 mutants, 36 hpf. |
|
Changes in Hh and Bmp signaling in glypican4fr6 mutants affect the expression of cxcl12a along the myoseptum. (A–C) Expression of cxcl12a in siblings (A) and glypican4fr6 mutants (B, C). In mutants gaps in cxcl12a expression persist until 48 hpf when the primordium migration is completed. (D–I) The Hh target gene patched2 becomes increasingly downregulated in glypican4fr6 mutants as the embryos develop. (J–U) On the other hand, the Bmp target gene id1 only shows signs of upregulation at around 18 hpf which becomes more severe by 38 hpf when the glypican4fr6 mutant primordia have completed their migration. |
|
glypican4fr6 mutant primordium cells migrate like wild type cells when transplanted into a wild type embryo. (A) Clone of glypican4fr6 mutant cells (Magenta+/GFP+) in a wild type primordium shows that mutant cells properly migrate toward the tail tip suggesting that the wild type environment rescued them. (B) Wild type primordium clone (Magenta+/GFP+) in a glypican4fr6 mutant environment. The wild type cells become rounder and they do not migrate posteriorly confirming a non-cell autonomous effect of glypican4. White arrows point to rosette constrictions in the primordium. See Movie S4. Anterior is to the left, and posterior is to the right. |
|
trilobite/vangl2 mutant primordia migrate all the way to the tail and express cxcl12a similarly to glypican4fr6 No-turning mutants. (A-B) Sibling and trilobite/vangl2 mutant embryos in the Tg(cldnb:lynGFP) background showing primordium migration by 48 hpf. trilobite/vangl2 mutant primordia migrate to the end of the tail, similar to their siblings and the glypican4fr6 No-turning mutants. (C–D'') Expression of the chemokine ligand cxcl12a in siblings (C) and trilobite/vangl2 mutant embryos (D). (C′–D′) Sibling and glypican4fr6 mutant primordia migrating on top of the cxcl12a-expressing horizontal myoseptum. (C''-D'') Same images as in C′ and D′ but focused on the cxcl12a expression in the horizontal myoseptum. |
Reprinted from Developmental Biology, 419(2), Venero Galanternik M., Lush, M.E., Piotrowski, T., Glypican4 Modulates Lateral Line Collective Cell Migration Non Cell-Autonomously, 321-335, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.