- Title
-
SPOP mutation leads to genomic instability in prostate cancer
- Authors
- Boysen, G., Barbieri, C.E., Prandi, D., Blattner, M., Chae, S.S., Dahija, A., Nataraj, S., Huang, D., Marotz, C., Xu, L., Huang, J., Lecca, P., Chhangawala, S., Liu, D., Zhou, P., Sboner, A., de Bono, J.S., Demichelis, F., Houvras, Y., Rubin, M.A.
- Source
- Full text @ Elife
|
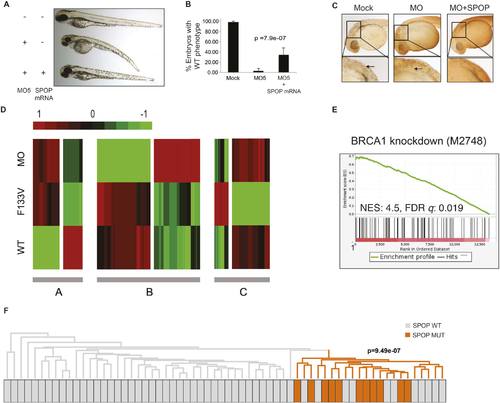
SPOP mediates DNA damage repair. (A) Evaluation of SPOP function during zebrafish development. Phenotype of morpholino-mediated Spop knockdown (MO5) in zebrafish embryos at 70 hr post fertilization (hpf). Injection of human SPOP mRNA (250 pg) rescued the phenotype. (B) Quantification of the rescue of SPOP phenotype after ectopic expression of human SPOP mRNA. Results are represented as s.e.m. (C) Whole mount TUNEL assay to determine apoptosis in zebrafish embryos. Arrows point to apoptotic cells (brown). Shown are representative images. (D) Heatmap representation of gene expression differences in zebrafish embryos ectopically expressing SPOP-wt or SPOP-F133V compared to SPOP knockdown by morpholino (MO). The list of genes can be found in Figure 2—figure supplement 3. Number of genes per block: A (198), B (429), C (223). (E) Gene set enrichment analysis (GSEA) of RNA sequencing data derived from zebrafish embryos expressing SPOP-wt or SPOP-F133V (24 hpf). Enrichment plot for the BRCA1 gene signature is shown. Molecular Signatures Database (MSigDB) systematic name indicated in brackets. (NES) Normalized Enrichment Score. (FDR) False Discovery Rate. (F) Dendrogram of human primary prostate cancer cases based on BRCA1 knockdown genes (MSigDB: M2748). Unsupervised clustering of RNA-seq data from human primary prostate cancer with wild-type (n = 53) or mutant SPOP (n = 11), performed on the BRCA1 knockdown gene signature (M2748) identified in zebrafish embryos by GSEA as shown in (E). |
|
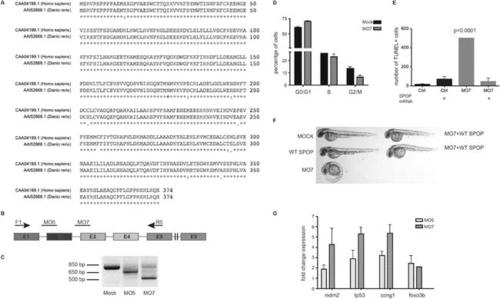
Spop knockdown in zebrafish using morpholinos results in a developmental phenotype. (A) Multiple protein sequence alignment of SPOP from Homo sapiens and zebrafish (Danio rerio) reveals 97.3% identity between humans and zebrafish on the amino acid level. (B) Schematic representation of zebrafish SPOP mRNA. Colored boxes indicate exons. MO5 and MO7 indicate the targeted sites of the SPOP-specific morpholinos. F1 and R5 indicate the sites were the primers bind, which were used to amplify exons 1–5 for molecular validation of morpholino efficiency. (C) Image of an agarose gel used to analyze zebrafish SPOP splice variants after morpholino treatment. (D) Propidium iodide-based cell cycle analyses of cells from zebrafish embryos (24 hr) treated with or without SPOP-specific morpholino. (E) Quantification of TUNEL-positive cells in the midbrain area of zebrafish embryos (24 hpf) treated with SPOP morpholino or in combination with WT-SPOP mRNA as shown in Figure 2C. (F) Example of zebrafish embryos (48 hpf) injected with a second splice-blocking morpholino (MO7). Rescue of the effects of MO7 on zebrafish development by coinjecting WT-SPOP mRNA. (G) qPCR-based validation of differentially expressed transcripts identified by RNA-seq. PHENOTYPE:
|