- Title
-
Protein-Trap Insertional Mutagenesis Uncovers New Genes Involved in Zebrafish Skin Development, Including a Neuregulin 2a-Based ErbB Signaling Pathway Required during Median Fin Fold Morphogenesis
- Authors
- Westcot, S.E., Hatzold, J., Urban, M.D., Richetti, S.K., Skuster, K.J., Harm, R.M., Lopez Cervera, R., Umemoto, N., McNulty, M.S., Clark, K.J., Hammerschmidt, M., Ekker, S.C.
- Source
- Full text @ PLoS One
|
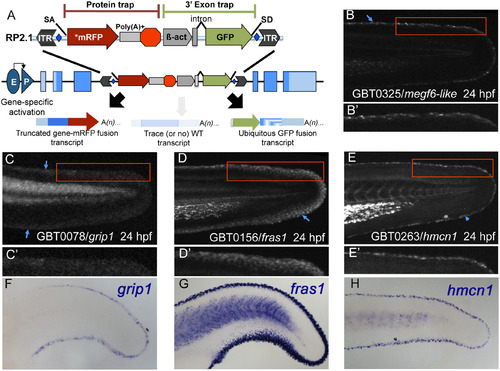
Gene-break transposon-based protein trapping identifies known and new epidermal median fin fold loci. (A) A schematic of the RP2.1 gene-break transposon (GBT) vector used in this study. Gene-breaking activity occurs when an endogenous locus with a GBT insertion is transcribed. The vector-supplied splice acceptor (SA) in the 5′ protein trap cassette intercepts the endogenous splicing machinery and transcript, redirecting them to read directly into an AUG-free mRFP sequence (*mRFP). That event generates a fusion transcript by tagging the 5′ portion of the endogenous transcript with mRFP. When translated, the mRFP fusion transcript will produce a potentially mutagenic truncated protein product. Simultaneously, the 3′ exon trap cassette uses the vector-supplied splice donor (SD) to create a GFP fusion transcript with the remaining downstream endogenous transcript. GBT alleles are revertible because loxP sites (blue diamonds) flank the cassettes, allowing the mutagenic elements to be excised in the presence of Cre recombinase. (B-E′) At 24 hours post-fertilization (24 hpf), GBT-generated mRFP fusion proteins from megf6amn0325Gt (B), grip1mn0078Gt (C), fras1mn0156Gt (D), and hmcn1mn0263Gt (E) localize along MFF edges. (B′, E′) Both megf6amn0325Gt (B′) and hmcn1mn0263Gt (E′) localize within a narrow region along the MFF edge (blue arrowheads). (C′, D′) grip1mn0078Gt (C′) and fras1mn0156Gt (D′) localization also follows the fin fold edge (blue arrowheads), though they are distributed somewhat more diffusely than are megf6amn0325Gt (B′) and hmcn1mn0263Gt (E′). (F-H) Whole-mount in situ hybridization (WISH) in 24 hpf wild-type embryos reveals similar MFF expression patterns of endogenous grip1 (F), fras1 (G), and hmcn1 (H) genes. The mRFP fusion protein localization patterns observed in the respective GBT lines recapitulate endogenous gene expression (C, D, E). EXPRESSION / LABELING:
|
|
GBT protein trapping identifies integument loci with epidermal or fin mesenchymal expression. As identified by mRFP localization, seven GBT alleles have unique epidermal expression patterns emphasizing epidermal continuity over the body and larval fin folds. They are: (A, B) nrg2amn0237Gt, (C, D) col4a4mn0275Gt, (E, F) arhgef25bmn0175Gt, (G, H) ahnakmn0196Gt, (I, J) capn12mn0261Gt, (K, L) fkbp10bmn0316Gt, and (M, N) msxcmn0245Gt. (B, D, I, J, K) Four lines, nrg2amn0237Gt, col4a4mn0275Gt, capn12mn0261Gt, and fkbp10bmn0316Gt, have “holes” or “gaps” in their epidermal pattern (green arrowheads) where neuromasts embedded in the basal layer exclude the mRFP-positive basal keratinocytes. (A, G, I, K, M) nrg2amn0237Gt (A), ahnakmn0196Gt (G), capn12mn0261Gt (I), fkbp10bmn0316Gt (K), and msxcmn0245Gt (M) are also expressed in the pectoral fin folds. (C-D, J, L, M-N) col4a4mn0275Gt, capn12mn0261Gt, fkbp10bmn0316Gt, and msxcmn0245Gt epidermal expression (blue arrowheads) can be difficult to discern among other expression pattern components. (I-N) capn12mn0261Gt, megf6amn0316Gt, and msxcmn0245Gt also show expression in fin mesenchymal cells (pink arrowheads). (C, D, K, L, M, N) Several lines are also expressed in other tissues. (C, D) col4a4mn0275Gt appears in myotomes and the vascular plexus. (K, L) fkbp10bmn0316Gt is seen in the notochord. (M, N) msxcmn0245Gt is strongly expressed in the brain, spinal cord, and sensory maculae. hpf, hours post-fertilization. Comparisons of the mRFP localization patterns with the expression pattern of the endogenous genes are shown in Supporting Information (S1 Fig) EXPRESSION / LABELING:
|
|
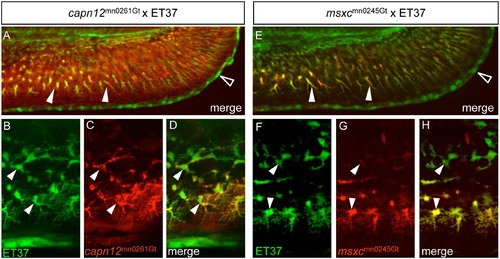
capn12 and msxc transgenes are co-expressed with a fin mesenchymal cell marker. (A, E) When capn12mn0261Gt and msxcmn0245Gt are crossed into an ET37 background, both Capn12mn0261Gt-mRFP and MsxCmn0245Gt-mRFP co-localize with ET37 EGFP-positive fin mesenchymal cells (FMCs) (arrowheads). ET37 also labels the cleft cells along the MFF edge (open arrowheads). Neither Capn12mn0261Gt-mRFP nor MsxCmn0245Gt-mRFP localizes to the cleft cells. (B-H) ET37 EGFP labels all FMCs within the fin (arrowheads), both proximal (upper arrowheads) and distal (lower arrowheads), and is present throughout FMC cell bodies and in the extended “arbors” of distal FMCs (lower arrowheads). Both Capn12mn0261Gt-mRFP and MsxCmn0245Gt-mRFP localization show similarities with and differences from ET37 EGFP. (B-D) Both ET37 EGFP and Capn12mn0261Gt-mRFP are expressed in proximal and distal FMCs (arrowheads) but unlike ET37 EGFP, Capn12mn0261Gt-mRFP localizes to the periphery of FMCs (arrowheads). (F-H) MsxCmn0245Gt-mRFP co-localizes with ET37 EGFP, but differs from Capn12mn0261Gt-mRFP. MsxCmn0245Gt-mRFP is much weaker in proximal FMCs (upper arrowheads) than in distal FMCs (lower arrowheads). Unlike Capn12mn0261Gt-mRFP, MsxCmn0245Gt-mRFP subcellular localization is not noticeably distinct from that of ET37 EGFP. EXPRESSION / LABELING:
|
|
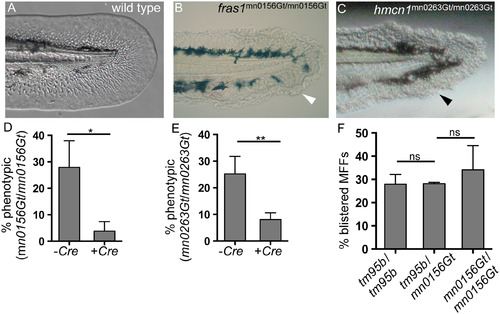
GBT protein trapping generates novel revertible alleles of known MFF loci. (A) By 48 hpf, wild-type MFFs are thin and flat, and the MFF edge appears smooth and regular. (B-C) Two ZIP gene-break alleles, fras1mn0156Gt and hmcn1mn0263Gt have homozygous recessive phenotypes, each of which results in blistered MFFs (arrowheads). (D-E) Both fras1mn0156Gt and hmcn1mn0263Gt behaved as classic revertible GBT mutant alleles in Cre reversion experiments. With both alleles, significantly fewer Cre mRNA-injected embryos were phenotypic, compared with their respective uninjected siblings. (D) For offspring of fras1mn0156Gt heterozygote incrosses, 27% (n = 156) of uninjected embryos (-Cre) were phenotypic; only 5% (n = 140) of Cre mRNA-injected siblings (+Cre) were phenotypic (p < 0.05). (E) For offspring of hmcn1mn0263Gt heterozygote incrosses, 25% (n = 146) of uninjected embryos (-Cre) were phenotypic; only 8% (n = 233) of Cre-injected siblings (+Cre) were phenotypic (p < 0.005). (F) The fras1mn0156Gt GBT allele fails to complement the pif tm95b ENU allele of fras1. Crossing pif tm95b heterozygotes with fras1mn0156Gt heterozygotes does not reduce the proportion of phenotypic offspring (28%, n = 445) (tm95b/mn0156Gt trans-heterozygotes) compared to either pif tm95b/tm95b (tm95b/tm95b, n = 332) or fras1mn0156Gt/mn0156Gt (mn0156Gt/mn0156Gt, n = 206) homozygotes. Percentages represent the mean of means (MOM); error bars represent standard deviations (SD). PHENOTYPE:
|
|
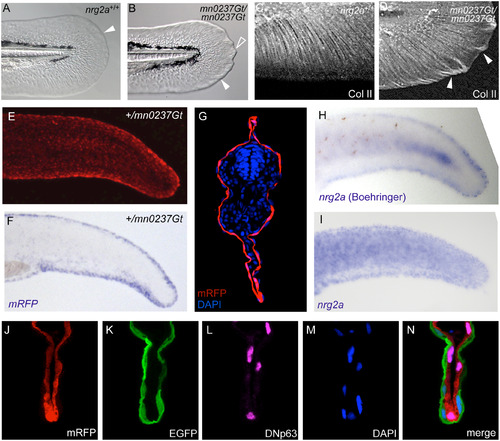
nrg2a mutants display altered MFF morphology, consistent with the epidermal localization of the Nrg2-mRFP fusion protein and of endogenous nrg2a transcripts. (A, B) By 48 hpf, nrg2amn0237Gt/mn0237Gt mutants (mn0237Gt/mn0237Gt) show altered MFF morphology. (A) Wild-type MFF edges are thin, flat, and continuously curved (arrowhead). (B) mn0237Gt/mn0237Gt mutant MFFs have thickened edges (arrowhead), and one or more pointed protrusions (open arrowhead). (C, D) Collagen II (Col II) immunostaining of actinotrichia support fibers within the MFF shows aberrant collagen accumulation and ectopic actinotrichia within mn0237Gt/mn0237Gt mutant apical MFFs (arrowheads) at 48 hpf. (E, G) At 24 hpf, Nrg2a-mRFP fusion protein is present in MFFs of heterozygous (+/mn0237Gt) embryos (E; view on tail of whole mount) and, at slightly lower levels, throughout the entire epidermis (G; section through tail region; immunostained for RFP and counterstained with DAPI). (F, H, I) Whole-mount in situ hybridization (WISH) demonstrates strong MFF expression of the GBT-generated fusion transcript (mRFP; F) in a representative +/mn0237Gt embryo at 24 hpf. When developed with Boehringer Blocking Reagent, WISH staining for endogenous nrg2a transcripts in 24 hpf wild-type embryos also revealed strong MFF expression of the endogenous gene (H, Boeringer). When developed without Boehringer Blocking Reagent, WISH staining further reveals uniform expression of the endogenous nrg2a gene throughout the entire epidermis (I). For cross-sections, see Honjo et al. (2008), Fig 6C [51]). (J-N) Co-labeling of a transverse section through the dorsal MFF of a +/mn0237Gt embryo at 24 hpf reveals restricted localization of the Nrg2a-mRFP fusion protein (J) in &916;Np63-positive basal keratinocytes (L), whereas the outer enveloping layer, labeled with EGFP (K), lacks the Nrg2a-mRFP protein; (M) DAPI counterstain; (N) merged image of different channels shown in (J-M). |
|
Usage of alternative first exons of nrg2a gene leads to differential cytosolic or nuclear localization of resulting protein isoforms. (A) A schematic of the nrg2a locus on linkage group 21 (LG21), including alternative first exons 1A, 1B and 1C. The GBT insertion is located in the intron separating exon 1C and exon 2. (B) RT-PCR analyses of 48 hpf sibling embryos from heterozygote (+/mn0237Gt) intercross, revealing that endogenous nrg2a 1B and 1C transcripts are only expressed in heterozygous (+/mn0237Gt) and wild-type siblings (+/+). nrg2a 1B-mRFP and 1C-mRFP fusion transcripts are only expressed in homozygous mutants (mn0237Gt/mn0237Gt) and heterozygous siblings (+/mn0237Gt). (C-N) Confocal images of MFF epidermis at 24 hpf detecting Nrg2a-RFP fusion protein (C, G, K; red), cell membranes (D, H, L; labeled with EGFP (green) after injection of egfp-caax mRNA at 1-cell stage), and cell nuclei (E, I, M; labeled with BFP (blue) after injection of nls-BFP mRNA at 1-cell stage). Panels (F, J, N) show merged images. (C-F) +/mn0237Gt embryo displays Nrg2amn0237Gt-RFP fusion protein both in the cytoplasm (empty arrowheads) and in the nuclei (filled arrowheads). (G-J) Wild-type embryo injected with mRNA encoding exon 1B-version of the Nrg2a-RFP fusion protein; the fusion protein is largely absent from the nucleus, but present in cyptoplasmic compartments. (K-N) Wild-type embryo injected with mRNA encoding exon 1C-version of the Nrg2a-RFP fusion protein; the fusion protein is present in the cytoplasm and the nuclei, similar to the distribution of the transgene-encoded protein (C-F). EXPRESSION / LABELING:
|
|
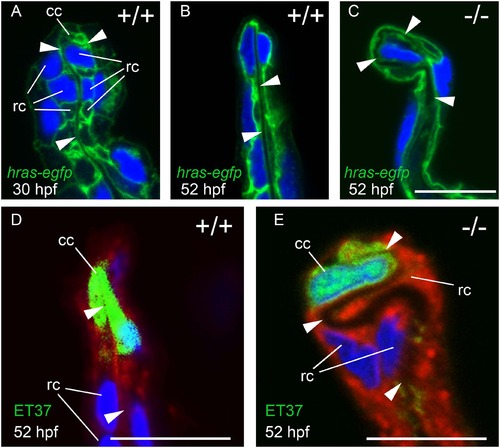
MFF cleft cells of nrg2a mutants are largely unaffected, but ridge cells display altered morphology. (A-C) Transverse sections through MFFs of Tg(Ola.Actb:Hsa.hras-egfp)vu119-expressing (hras-egfp) wild-type and nrg2a mutant embryos. Green: membrane-bound EGFP, blue: DAPI. (D-E) ET37 EGFP is expressed in MFF cleft cells (cc). ET37-EGFP: green, CellMask: red, DAPI: blue. (A) At 30 hpf, wild-type ridge cells (rc) are roughly cuboidal, with parallel apical and basal domains. The dermal space (ds) has not yet straightened, especially within the apical MFF terminus bounded by the cleft cell (cc). (B, D) By 52 hpf, the dermal space of wild-type embryos has straightened (arrowheads) and has invaginated into the basal side of the cleft cell. Ridge cells have elongated laterally, adopting a flat, planar, epithelial morphology. Their apical and basal domains are essentially parallel. (C, E) In nrg2a mutants, the cleft cell (cc) is present and contains the basal invagination of the dermal space (cleft; E) as in wild-type siblings. In contrast, nrg2a mutant ridge cells have elongated incorrectly and display an abnormal morphology, bulging basally into the dermal space, which acquires a serpentine-like appearance. Scale bar: 10 µm. Abbreviations: cc, cleft cell; rc, ridge cell; arrowheads point to dermal space. |
|
MFF ridge cell in nrg2a mutants display alterations in basolateral versus apical dimensions. (A-H) Transmission electron micrographs (TEM) of the distal-most region within wild-type and nrg2a mutant MFFs at 36 hpf (A, B, E-H) or 52 hpf (C,D). (A) By 36 hpf, wild-type ridge cells (arrowhead) have begun elongating laterally. Their apical and basal surfaces are parallel to each other and to the dermal space. (C) By 52 hpf, wild-ridge cells have continued elongating and have maintained their arrangements. (B, D) In nrg2a mutants (mn0237Gt/mn0237Gt), ridge cells (arrowheads) are morphologically distorted, fail to stay aligned parallel to the direction of the fin, and bulge into the dermal space, giving it a serpentine-like appearance. (E-H) Relative basolateral and apical dimensions in nrg2a mutants are distorted compared to their wild-type counterparts. To illustrate the changes, identical images are shown side by side with and without marked ridge cell borders. (E, F) By 36 hpf, apical (red) and basal (blue) borders of wild-type ridge cells are roughly parallel and of comparable lengths; lateral borders with neighboring basal keratinocytes are in white. (G, H) An example of mn0237Gt/mn0237Gt mutant ridge cells bulging into the dermal space. The pictured bulge consists of two adjacent ridge cells sharing an exaggeratedly lengthened lateral border (white) and with enlarged basal (blue) borders, but strongly reduced apical borders (red). Scale bars: 2 µm. Abbreviations: ds, dermal space; e, EV; cc, cleft cell; arrowheads point to ridge cells. |
|
The nrg2a MFF phenotype can be mimicked and synergistically enhanced via chemical ErbB inhibition, and is characterized by reduced pAKT levels. (A-J) Pharmacological inhibition of ErbB signaling during MFF morphogenesis (24 through 52 hpf) induces nrg2amn0237Gt/mn0237Gt-like effects in wild-type and, with even higher frequencies, nrg2a+/mn0237Gt heterozygous embryos. (A) MFFs of wild-type embryos treated both with a low dose (1 µM) (28%; n = 120; p = 0.005) or a high dose (20 µM) (76%; n = 102; p < 0.0001) of PD168393 show an nrg2amn0237Gt/mn0237Gt-like MFF morphology at 52 hpf. (B) nrg2a+/mn0237Gt heterozygous embryos (red) are significantly more sensitive to low-dose (1 µM) PD168393 treatment, and display nrg2amn0237Gt/mn0237Gt-like MFF morphology with a higher frequency (64%; n = 152) than treated nrg2a+/+ wild-type siblings (blue) (n = 23%; n = 116; p < 0.0001) or treated unrelated wild-type embryos (grey) (28%; n = 120; p < 0.0001). (D-H) Live images of MFFs of representative examples of wild-type (C, E, G) or nrg2a+/mn0237Gt heterozygous (D, F, H) embryos at 52 hpf, after treatment with DMSO (control; C, D), 1 µM PD168393 (E, F) or 20 µM PD168393 (G, H). (I, J) TEM transverse sections of the apical MFF reveal a ridge cell phenotype in PD168393-treated wild-type embryos at 52 hpf. (I) An untreated wild-type embryo has correctly elongated ridge cells (red arrowhead) and a straight dermal space (ds). (J) A sibling embryo treated with 20 µM PD168393 displays basal bulging of MFF ridge cells towards the center the fin fold (red arrowheads) and a corresponding serpentine-like folding of the dermal space (ds), resembling the defects of the nrg2a mutant (compare with Fig 8D). (K, L) Whole-mount in situ hybridization (WISH) for egfra (K) and erbb3a (L) in wild-type embryos at 34 hpf (lateral views of tail) reveals epidermally expressed erbb3a transcripts (L), whereas egfra transcipts are absent in the epidermis, but present in the somites (K). (M-P) Anti-pAKT immunofluorescence of wild-type (M, N) and nrg2a mutant (O, P) embryo at 34 hpf; transverse sections through MER region of MFF, counterstained with DAPI (N, P). The wild-type embryo (M, N) displays pAKT localization in distal epidermal MFF cells (ridge cells and cleft cells; arrowheads), whereas pAKT levels in more proximal epidermal MFF cells are much lower. In addition, pAKT is localized at the tight junctions of the outer EVL (arrows), consistent with previously described roles of pAKT to phosphorylate tight junction proteins ZO-1 and Occludin, and to tighten the junctions [122]. In the nrg2a mutant (O, P), pAKT signals in ridge and cleft cells are strongly reduced (arrowheads), while pAKT signals at EVL tight junctions are unaltered (arrows). |
|
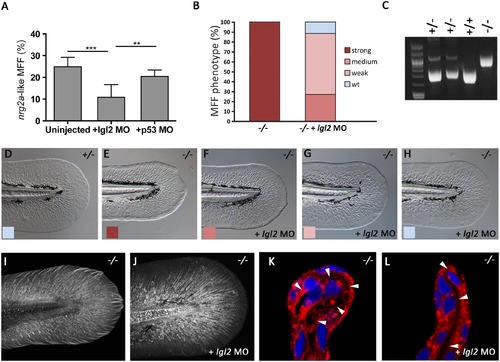
The MFF phenotype of nrg2a mutants is rescued upon concomitant loss of Lgl2 function. (A) At 52 hpf, morpholino (MO)-mediated knockdown of lgl2 significantly ameliorated the nrg2a mutant phenotype (12%, n = 639, p < 0.0001) relative to uninjected embryos of an nrg2a -/+ intercross (25%, n = 943). Percentages represent the mean of means (MOM); error bars represent the standard deviations (SD). (B) Percentages of genotyped nrg2a-/- mutants with a strong, medium, weak, or wild-type MFF phenotype, classified by morphological criteria at 52 hpf. While uninjected nrg2a mutants (n = 20) all display a strong phenotype, lgl2 MO-injected mutants (n = 26) show medium, weak or no MFF defects. (C) PCR products obtained via nrg2a genotyping of representative nrg2a -/+, +/+ and-/- embryos at 52 hpf (see Materials and Methods). (D-H) Tail fins of representative live embryos at 52 hpf, as used for quantitative classification in panel B: wild-type (D), uninjected nrg2a-/- mutant with strong MFF phenotype (E), and lgl2 MO-injected nrg2a -/- mutant embryos with medium (F), weak (G) or wild-type (H) phenotype. (I, J) Tail fins of genotyped uninjected (I) and lgl2 MO-injected (J) nrg2a -/- mutant embryo at 48 hpf. Col II immunostaining reveals a normalized organization of collagenous actinotrichia within the dermal space of the Nrg2a/Lgl2-double-deficient embryo (J; compare with Fig 5C for wild-type condition). (K, L) Transverse sections through the dorsal MFF of a genotyped uninjected (K) and an lgl2 MO-injected (L) nrg2a -/- mutant embryo at 52 hpf; CellMask (red) and DAPI (blue) staining reveals a rescue of the dermal space (indicated by arrowheads) from a serpentine-like organization (K) to a straight organization (L) in the Nrg2a/Lgl2-double-deficient embryo (L; compare with Fig 7D for wild-type condition). |
|
mRFP fusion protein distribution in GBT lines recapitulates the endogenous expression patterns of arhgef25b, fkbp10b, and msxc. Panels (A, A′, C, C′, E, E′) show the same in vivo fluorescence images of mRFP localization in GBT lines arhgef25bmn0175Gt (A, A′), fkbp10bmn0316Gt (C, C′) and msxcmn0245Gt (E, E′) as in Fig 2E, 2F, 2K, 2L, 2M, and 2N, respectively. Panels underneath show images of wild-type embryos of comparable stages (indicated in hpf) and orientations following whole-mount in situ hybridization (WISH) with arhgef25b (B, B′), fkbp10b (D, D′), or msxc (F, F′) RNA probes. mRFP localization in the epidermis covering the yolk in the arhgef25bmn0175Gt gene-break allele (A) corresponds to endogenous arhgef25b gene expression (B). Both arhgef25bmn0175Gt mRFP localization and endogenous arhgef25b gene expression are also observed in the MFF (A′, B′). Furthermore, just as the gene-break alleles fkbp10bmn0316Gt (C, C′) and msxcmn0245Gt (E, E′) show mRFP localization in fin mesenchymal cells (FMCs), fkbp10b and msxc genes show endogenous FMC expression (C-F′; also compare with MsxCmn0245Gt-mRFP fusion protein localization in Fig 3E–3H). mRFP localization to the pectoral fin buds observed in the fkbp10bmn0316Gt allele (C) parallels endogenous fkbp10b expression in the pectoral fin buds (D). fkbp10b also shows expression in the posterior region of the notochord (D′). The weaker WISH signal in anterior (earlier specified) regions of the notochord compared to the mRFP signal (C′) points to transient expression of the endogenous gene in notochord cells, and higher stability of the GBT-generated mRFP fusion protein than the endogenous transcript. The endogenous msxc gene is also expressed in the maculae of the inner ear and in the pectoral fin buds (F), confirming that the msxcmn0245Gt GBT allele and its resulting mRFP fusion protein also recapitulate endogenous expression in tissues other than the skin (E). |
|
nrg2a mutant larvae have altered pectoral fin folds, lack swim bladders and die between 8 and 12 dpf. (A-D) Images of live larvae at 7 dpf. (A) Wild-type pectoral fin folds (PFF) typically follow a continuous arc such that the PFF edge lies close to the body when larvae are at rest. (B) PFFs in nrg2a mutant larvae (mn0237Gt/mn0237Gt) often depart from a continuous arc shape and/or have thickened edges similar to the MFF phenotype (arrowhead). In addition, wild-type larvae have well-developed swim bladders (C), but swim bladders fail to develop in mn0237Gt/mn0237Gt mutant larvae (D, arrowhead). (E) Graphical illustration of survival rates of mn0237Gt/mn0237Gt mutant larvae (n = 104 larvae per class). |
|
Cleft cells in nrg2a mutants are largely unaffected, whereas ridge cells display expanded basal and reduced apical domains. Transmission electron micrographs (TEM) of distal-most region of dorsal MFF of wild-type (A) and nrg2a mutant (mn0237Gt/mn0237Gt) (B-D) embryos at 36 hpf (A, B) or 52 hpf (C, D). (A, B) Cleft cell (cc) morphogenesis creates the cleft, an invagination of the nascent dermal space (ds, red arrow) into the cleft cell. White lines trace cleft cell boundaries; red arrow termini (red arrowheads) indicate termination of the dermal space within the cleft. The nrg2a mutant (B) has an intact cleft cell with normal morphology. (C, D) Representative example of a ridge bulging into the dermal space, consisting of a single ridge cell with an extended basal border (blue; D) and a noticeably reduced apical border (red, D). Lateral borders are in white (D). For clarity, identical images are shown side by side with (D) and without (C) marked ridge cell borders. Magnification: 10,000X, scale bar: 2 µm. (A-D) 36 hpf; (E-F) 2 dpf. Abbreviatiations: cc, cleft cell; ds, dermal space; e, EVL cell; rc, ridge cell. |
|
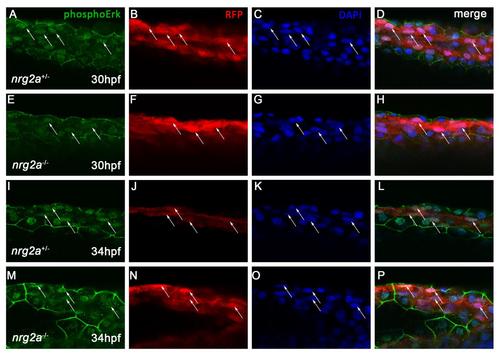
pERK levels in nrg2a mutant MFF basal keratinocytes are unchanged. Confocal images of whole-mount dorsal MFFs from nrg2a+/mn037Gt (A-D, I-K) and nrg2amn037Gt/mn037Gt (E-H, M-P) embryos at 30hpf and 34 hpf do not reveal changes in activated ERK (phosphoERK, pERK) levels in ridge cells. Embryos were immunostained for phosphoErk (A, E, I, M) and basal keratinocytes were immunostained for Nrg2a-mRFP (B, F, J, N). Nuclei were counterstained with DAPI (C, G, K, O). Merged images are shown in D, H, K, and M. Arrows indicate ridge cells that are positive for pERK both in nrg2a+/mn037Gt and nrg2amn037Gt/mn037Gt embryos. |