- Title
-
Isolation of Novel CreERT2-Driver Lines in Zebrafish Using an Unbiased Gene Trap Approach
- Authors
- Jungke, P., Hammer, J., Hans, S., Brand, M.
- Source
- Full text @ PLoS One
|
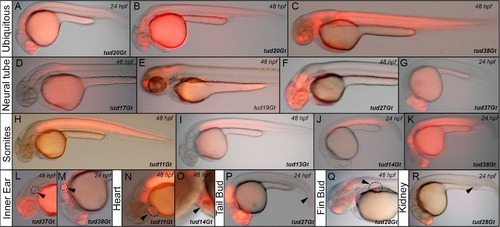
Native mCherry expression of CreERT2-driver lines in a variety of embryonic tissues. (A-C) Ubiquitous mCherry expression in tud20Gt at 24 and 48 hpf, and tud38Gt at 48 hpf. (D-G) CreERT2-driver lines expressing mCherry in restricted patterns of the neural tube: (D) hindbrain and spinal cord expression in tud17Gt at 48 hpf, (E-G) fore-, mid- and hindbrain expression in tud19Gt and tud27Gt at 48 hpf and fore- and midbrain expression in tud37Gt at 24 hpf. (H-K) Somitic mCherry expression in tud11Gt and tud13Gt at 48 hpf as well as in tud14Gt and tud38Gt at 24 hpf. (L, M) mCherry expression in the developing inner ear in tud37Gt at 48 hpf and tud38Gt at 24 hpf. (N, O) CreERT2-driver lines expressing mCherry in the embryonic heart in tud11Gt and tud14Gt at 48 hpf. (P) mCherry expression in the tail bud in tud27Gt at 24 hpf. (Q) Fin bud expression in tud28Gt at 48 hpf. (R) CreERT2-driver line tud28Gt shows mCherry expression in the kidney anlagen at 24 hpf; Bold letters indicate CreERT2-driver lines with known gene trap integrations. |
|
Identification of functional CreERT2-driver lines. (A) Scheme of the embryonic functionality assay. (B) 27 CreERT2-driver lines are shown with respect to CreERT2 expression at 24 hpf (CreERT2) and the respective embryonic functionality assay indicated by native EGFP fluorescence. EXPRESSION / LABELING:
|
|
Functionality of CreERT2-driver lines at larval stages. tud29Gt, tud37Gt and tud38Gt are shown with respect to CreERT2 expression at 48 hpf (CreERT2) and EGFP fluorescence in the presence and absence of 4-hydroxy-Tamoxifen (4-OHT) at 100 hpf. EXPRESSION / LABELING:
|
|
CreERT2-driver lines expressing in the embryonic neural tube. (A, B) Pan-neural expression of CreERT2 revealed by in situ hybridization in tud20Gt and tud30Gt at 24 hpf. (C,D) Expression of CreERT2 in the spinal cord in tud17Gt at 24 and 48 hpf. (E,F) Hindbrain expression of CreERT2 in tud18Gt and tud35Gt at 24 hpf. (G-J) Fore-/Midbrain expression and (K-Q) other restricted patterns of CreERT2 in tud15Gt, tud19Gt, tud27Gt, tud28Gt, tud34Gt, tud35Gt and tud37Gt at 24 and 48 hpf, respectively. Bold letters indicate CreERT2-driver lines with known gene trap integrations. (See text for detailed description of expression patterns.); ep: epiphysis; fb: forebrain; hb: hindbrain; mb: midbrain; mhb: mid-hindbrain-boundary; t: tectum; tc: telencephalon. |
|
CreERT2-driver lines expressing in the embryonic eye. (A-F) Expression of CreERT2 in the developing retina in tud20Gt, tud32Gt and tud35Gt at 24 and 48 hpf, respectively. (G-L) CreERT2 is expressed in the developing lens in tud15Gt, tud27Gt and tud34Gt at 24 and 48 hpf, respectively. White dotted circles mark the retina (A-F) and lens (G-H). Bold letters indicate CreERT2-driver lines with known gene trap integrations. (See text for detailed description of expression patterns.) |
|
CreERT2-driver lines expressing in the somites. Expression of CreERT2 in (A-D) tud11Gt, (E-H) tud13Gt, (I-L) tud14Gt and (M-P) tud38Gt at 24 and 48 hpf.; Bold letters indicate CreERT2-driver lines with known gene trap integration. (See text for detailed description of expression patterns.) as: anterior somites; psm: presomitic mesoderm. |
|
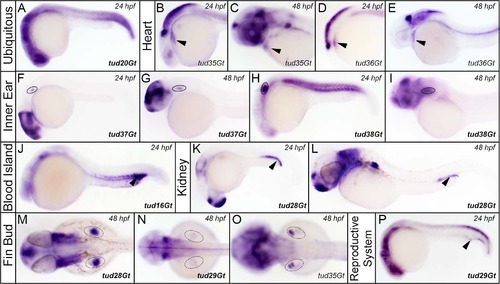
CreERT2-driver lines expressing in various embryonic tissues. (A) Ubiquitous expression of CreERT2 in tud20Gt at 24 hpf. (B-E) CreERT2 is expressed in the developing heart in tud35Gt and tud36Gt at 24 and 48 hpf. (F-P) Expression of CreERT2 can be detected in the anlagen of (F-I) the inner ear in tud37Gt and tud38Gt (J) blood island in tud16Gt, (K,L) kidney in tud28Gt (M-O) fin buds in tud28Gt, tud29Gt and tud35Gt and (P) the reproductive system in tud29Gt at 24 and 48 hpf. Bold letters indicate CreERT2-driver lines with known gene trap integrations. (See text for description of expression patterns.) |
|
Gene trap insertion into the otx1b locus of tud37Gt. (A) Schematic drawing of the otx1b locus comprising of four exons (E1-E4) encoding a homeobox domain transcription factor. White boxes represent the 5′ and 3′ untranslated regions separated by the open reading frame in pink. The mCT2aC-cassette integrated into E4. (B) Bright field images of homozygous mutant tud37Gt embryos and heterozygous siblings. In comparison to heterozygous siblings, homozygous mutant tud37Gt embryos show defects in the developing eye, fore-/midbrain, ear and heart (white arrowheads) as well as a bend body shape. PHENOTYPE:
|
|
Insertion of the mCT2aC gene trap vectors recapitulates the endogenous gene expression pattern. Comparison of the endogenous otx1b expression in wild-type embryos with the CreERT2 expression pattern in tud37Gt embryos which has been mapped to the otx1b locus at 24 and 48 hpf. fb: forebrain; mb: midbrain; ov: otic vesicle. |