- Title
-
An In Vivo Requirement for the Mediator Subunit Med14 in the Maintenance of Stem Cell Populations
- Authors
- Burrows, J.T., Pearson, B.J., Scott, I.C.
- Source
- Full text @ Stem Cell Reports
|
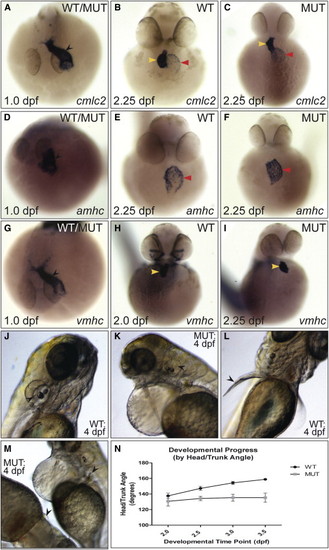
Characterization of the log Mutant Phenotype (A–I) Expression of cardiac markers in (WT) and log mutant (MUT) embryos. While at 1.0 dpf the linear heart tube (black arrowhead) was apparent in all embryos (“WT/MUT”), at 2.25 dpf log mutant hearts were unlooped, with the ventricle directly above the atrium (yellow and red arrowheads, respectively). (J–M) At 4.0 dpf, semicircular canals in the otic vesicles (black arrow heads in J and K) and elongation of the pectoral fins (black arrowheads in L and M) were absent in mutant embryos. (N) Change in head/trunk angle over time in WT and mutant embryos (10 WT and 20 mutants measured per time point). See also Figure S1. |
|
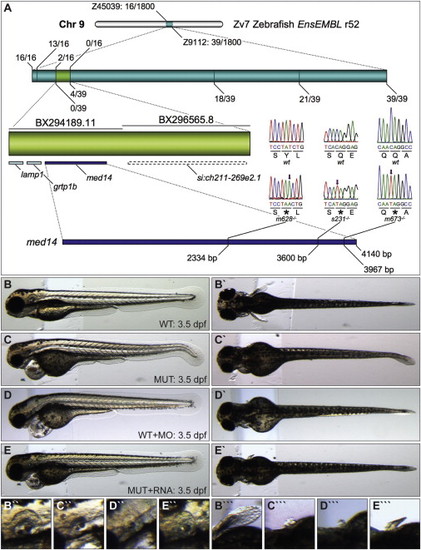
Logelei Results from a Mutation in med14 (A) Recombination frequency mapping results. Screening of 1,800 map-cross mutant embryos with markers Z45039 and Z9112 revealed 16 and 39 flanking recombinant embryos, respectively. The zero-recombinant region (light green, contained in two bacterial artificial chromosomes [BACs]) was refined with additional markers. Sequencing of med14 revealed a base pair substitution (arrow) leading to a premature stop codon () in each of the three log alleles. (B–E′′′) Lateral and dorsal views of the otic vesicle and pectoral fin of 3.5 dpf WT (B–B′′′), log mutant (C–C′′′), med14 morpholino-injected (D–D′′′), and log mutant injected with med14 RNA (E–E′′′) embryos. PHENOTYPE:
|
|
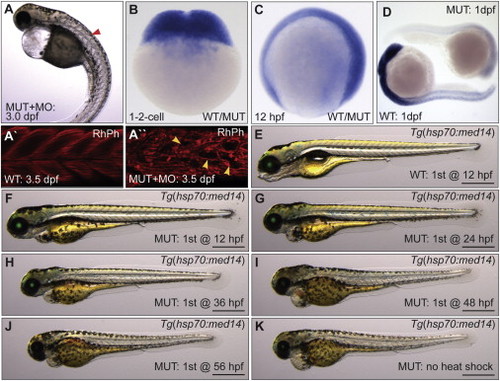
Expression and Timing Requirement of med14 for Development (A) Worsening of the log mutant phenotype by injection of morpholino (MO) targeting med14 (red arrowhead denotes somite defect). (A′ and A′′) Somite structure of 3.5 dpf rhodamine phalloidin (RhPh) stained WT and MO-injected mutant embryos. Defects in muscle fiber patterns of mutant embryos injected with MO are shown (yellow arrowheads). (B–D) RNA ISH analysis shows that med14 is broadly expressed at the one- and two-cell stage and at 12 hpf (“WT/MUT” denotes unknown genotype). At 24 hpf, broad med14 expression is undetectable in zygotic med14 mutants. (E–K) Temporal rescue of the log mutant phenotype using Tg(hsp70:med14, α-crystallin:EGFP), with initial heat shock performed at the specified time, and then every 12 hr following until 5 dpf. Scale bars, 0.5 mm. PHENOTYPE:
|
|
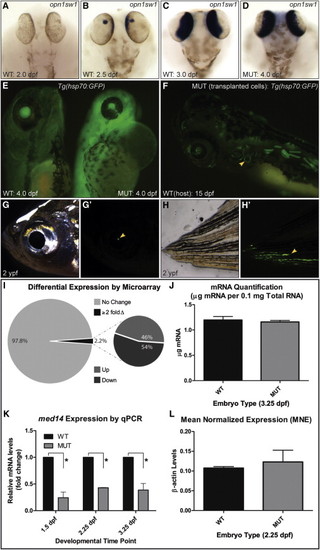
Transcription Is Not Largely Affected in log (med14) Mutants (A–D) Expression of opn1sw1, initiated by 2.5 dpf in WT embryos, reaches WT levels in log mutant embryos by 4.0 dpf. (E) Tg(hsp70:EGFP) log mutant embryos heat shocked at 3.5 dpf show robust GFP signal. (F–H′) log mutant cells transplanted into WT hosts (traced using a β-actin:EGFP transgene) are evident in 15 dpf hosts (including in the semicircular canals, yellow arrowhead in F) and survive until 2 ypf (years post-fertilization, yellow arrowheads). (I) Summary of results of microarray analysis on cDNA from 2.25 dpf WT versus log mutant embryos. (J) Quantification of mRNA per 0.1mg total RNA at 3.25 dpf from WT and log mutant embryos. (K) Normalized qPCR values for med14 expression in mutant relative to WT control embryos (p < 0.05 using the one-tailed unpaired Student’s t test). (L) Mean normalized expression (MNE) of β-actin in WT and MUT embryos at 2.25 dpf calculated using a universal reference approach. No significant difference (p = 0.63) was observed between WT (0.107 ± 0.00380) and log MUT (0.123 ± 0.0297) samples (n = 3) by one-tailed unpaired Student’s t test. For (J)–(L), three biological replicates of 450 (J) or 10 (K and L) embryos were used. |
|
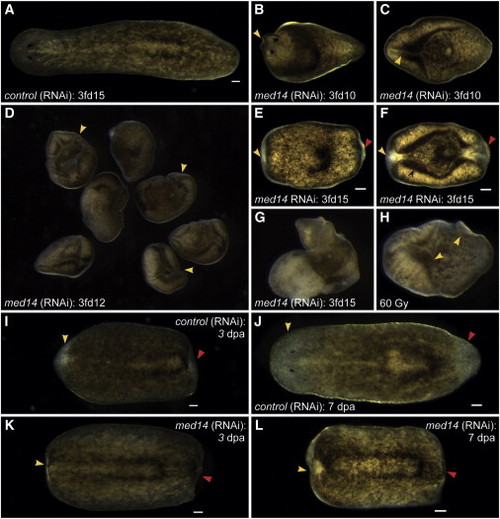
Smed-med14 Is Required for Tissue Homeostasis and Regeneration in Planarians (A–F) med14(RNAi) animals show defects in homeostasis, with head regression (yellow arrowheads) and ventral curling apparent in 100% of cases by 3fd10. (G and H) By 3fd12–15, head and tail regression (yellow and red arrowheads) and the beginnings of lysis are apparent in 100% of med14(RNAi) animals, similar to what is observed following irradiation-mediated depletion of the stem cell pool. (I–L) In trunk fragments that are regenerating a head (yellow arrowhead) and a tail (red arrowhead), no regeneration is observed by 7 dpa in med14(RNAi) animals. Scale bars, 100 µm. |
|
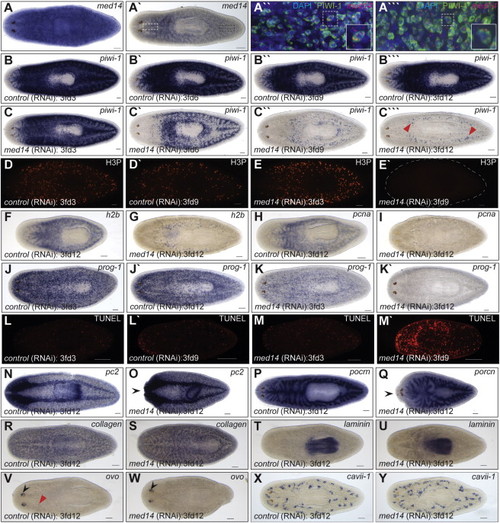
Smed-med14 Is Necessary for the Maintenance of Adult Stem Cells (A) ISH for med14 in wild-type intact animals showing ubiquitous staining. (A′) A stem cell like expression pattern is evident with reduced staining. (A′′ and A′′′) Confocal image at 25× magnification of med14 fluorescent RNA ISH (red) and PIWI antibody staining (green) in the planarian head (white dashed box in A2) and tail (black dashed box in A′) respectively. The boxed area in each is enlarged for clarification. med14 is expression in, but not limited to, the stem cell population. (B–C′′′) ISH analysis using a stem cell specific riboprobe (piwi-1) during a time course of med14(RNAi). By 3fd12, the stem cell population is largely absent in med14(RNAi) animals. The remaining piwi-1+ cells at 3fd12 (C′′′) may represent primordial germ cells (red arrowheads in M). (D–E′) Loss of proliferative phosphorylated histone H3 (H3P) +’ve cells in med14(RNAi) animals by 3fd9 (E′). (F–I) Expression of S-phase markers h2b and pcna in WT and med14 RNAi animals at 3fd12. (J–K′) By 3fd3, the progenitor cell population in med14(RNAi) animals (marked by prog-1 expression) is reduced compared with controls and completely absent by 3fd12. (L–M′) Increased cell death by 3fd9 as observed by whole-mount TUNEL analysis in med14(RNAi) animals. (N–Y) Normal expression of markers of differentiated cell types in med14(RNAi) animals as evident for the nervous system (pc2), gut (porcn), muscle (collagen), pharynx (laminin), eyes (ovo), and protonephridia (cavii-1). Head regression is evident in some treated worms (black arrow heads in O and Q). Eye progenitors (red arrow head in V) are not observed in med14(RNAi) animals despite ovo expression in the eye spots (black arrow heads in V and W). Scale bars, 100 µm. See also Figure S2. |
|
A Conserved Requirement for med14 in the Maintenance of Stem Cell Populations (A–D) Expression of retinal stem cell marker mz98 (arrowhead) is present, but reduced in log mutants at 2.25 dpf, and absent at 3.25 dpf. (E–H) Expression of the hematopoietic stem cell marker cmyb is initiated in 2.25 dpf log mutant embryos (arrowhead), but largely absent by 3.25 dpf. (I and J) Expression of the putative gut stem cell marker lgr4 is not observed in 3.25 dpf log mutant embryos (arrowhead, note expression in WT). (K and L) Robust tail fin regeneration in WT as compared with log mutant embryos at 4 dpf following amputation (at area of dotted line) at 2 dpf. (M–R) o-Dianisidane staining of red blood cells in trunks of WT and log mutant embryos. Scale bars, 0.5 mm. (S and T) Confocal projections of 2.5 dpf Tg(cmcl2:nlsDsRedExpress) WT and log morphant hearts. (U) Quantification of myocardial cell number in WT and morphant hearts shows a significant decrease in morphants (p = 0.0017, n = 6 for both conditions). (V) Cardiac edema and heart defects remain in 3.5 dpf Tg (cmlc2:med14, α-crystallin:EGFP) log mutant embryos. (W and X) Expression of the second heart field marker ltbp3 in the arterial pole of the heart (arrowhead) is reduced in log mutants at 48 hpf. EXPRESSION / LABELING:
|