- Title
-
Gap junctions composed of connexions 41.8 and 39.4 are essential for colour pattern formation in zebrafish
- Authors
- Irion, U., Frohnhöfer, H.G., Krauss, J., Çolak Champollion, T., Maischein, H., Geiger-Rudolph, S., Weiler, C., Nüsslein-Volhard, C.
- Source
- Full text @ Elife
|
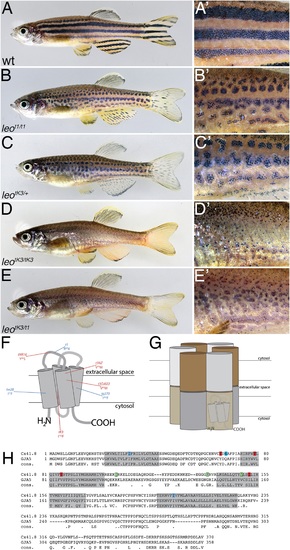
The leo mutant phenotype. Wild-type zebrafish (A) show a pattern of dark and light stripes on the body and on anal- and tail-fins. At higher magnification (A'), dark melanophores in the stripe regions and orange xanthophores in the light stripe regions are discernible. In mutants homozygous for leot1 (B) and heterozygous for leotK3 (C), the stripes are dissolved into spots. Clusters of melanophores are still visible (B' and C'). Fish homozygous for leotK3 (D) or trans-heterozygous for leotK3 over leot1 (E) show an identical phenotype of a completely dissolved pattern. Individual melanophores that hardly cluster together are still present, mostly associated with blue iridophores (D' and E'). In (F) a cartoon of Connexin 41.8 is depicted showing the positions of the leo mutations. Gap junctions are composed of two hemi-channels in adjacent cells. Each hemi-channel is made of six connexin subunits, they can be identical (homomeric) or different (heteromeric). In (G) a heteromeric/heterotypic gap junction is schematically shown. An alignment of the amino acid sequence from zebrafish Cx41.8 with its human orthologue, GJA5, is shown in (H). The transmembrane regions are shaded in grey. Newly identified point mutations in Connexin 41.8 are highlighted in red: leotNR16: Y66S; leotNZ: V79M; leot3OJ022: V85M; leotK3: I152F. Already known alleles are highlighted in blue: leotw28: I31F; leot1: R68X; leotq270: I202F. Polymorphisms found in sequences from wild-type fish are highlighted in green: 106 R/K, 136 G/R; 149 V/I. |
|
Dominant enhancers of the leot1 phenotype. The spotted pattern of fish homozygous for leot1 (A) is lost when they carry an additional mutant allele for luctXA9 (B) or luctXG1 (E). Fish heterozygous for luctXA9 (C) and luctXG1 (F) have undulating and disrupted stripes on their flanks. In mutants homozygous for luctXA9 (D) or luctXG1 (G) the pattern is almost as strongly disrupted as in leotK3 mutants (compare to Figure 1D). PHENOTYPE:
|
|
luc loss-of-function leads to patterning defects. A comparison of the amino acid sequences of Cx39.4 of zebrafish, encoded by the luc gene, and human Cx37 (GJA4) is shown in (A). The transmembrane domains are shaded in grey, the residues mutated in luctXA9 (T29 P) and luctXG1 (W47 L) are highlighted in red. Note the extension of the N-terminal cytoplasmic domain of Cx39.4 by two amino acid residues. In (B) the beginning of the coding sequence for Cx39.4 is shown, the mutations in luctXA9 and luctXG1 are indicated in red above the sequence; the CRISPR target site is underlined. The sequence traces of wild-type fish and two fish homozygous for deletions of 2 and 22 bp are shown in (C). The predicted amino acid sequences for these mutants are indicated. In (D) a fish homozygous for the 2 bp deletion is shown, the striped pattern on the flank of the fish is disrupted and partly dissolved into spots, the fins are almost normal. Double mutants for leot1 and luc loss-of-function (E) have only very few melanophores on the flank and show an almost uniform pattern of iridophores and xanthophores. PHENOTYPE:
|
|
The leo and luc phenotypes arise during metamorphosis. In wild-type dense iridophores of consecutively formed interstripes become visible during stages PR (A), J (F), and J++ (K). Melanophores appear in the stripe regions. In luc loss-of-function (B, G, L), leot1 (C, H, M), luctXG1 (D, I, N), and leotK3 mutants (E, J, O) iridophores of the first interstripe form normally at stage PR (B–E), however, the melanophores ventral to the first interstripe are more scattered. In the mutants at stage J (G–J) the second interstripe fails to form in the ventral region where dense iridophores appear in irregular positions. Later at stage J++ irregular stripes and spots become visible in luc loss-of-function and leot1 mutants (L and M), in luctXG1 and leotK3 mutants (N and O) the number of melanophores is low and dense iridophores cover increasing areas on the flank of the fish. Scale bars: 0.25 mm. PHENOTYPE:
|
|
Complete loss of the pigment pattern in double mutants of nac, pfe, and shd with leotK3. Mutants lacking one pigment cell type, nac (A and A′), pfe (C and C′), and shd (E and E′), only form remnants of the pigmentation pattern. In nac mutants fields of dense iridophores covered with compact xanthophores (corresponding to light stripes) and loose iridophores (corresponding to dark stripes) are discernible as patches (A′). In homozygous double mutants leotK3;nac (B) the regions of dense iridophores covered by compact xanthophores expand at the expense of blue iridophore areas. In pfe mutants (C) in the absence of xanthophores the melanophores form patches in the regions around the first light stripe but are increasingly more disorganized at some distance to it (C′). In double mutants of pfe with homozygous leotK3 melanophores are distributed almost evenly as individual cells over the flank of the fish (D). In shd mutants (E), where no iridophores are present, melanophores and xanthophores only partly form the first light and dark stripes (E′). In double homozygous double mutants leotK3;shd this residual pattern is lost and only very few individual melanophores are left on the flank of the fish (F). |
|
leo gap junctions are required in melanophores and xanthophores but not in iridophores. Typical examples of chimeric animals derived from blastomere transplantations are shown. Cells from leotK3;nac;pfe donor embryos, which can provide iridophores as their only chromatophore type, were transplanted in rse hosts, which lack most iridophores. The resulting chimeric animals (17 clones in 34 fish) show that mutant iridophores form a wild-type pattern when confronted with wild-type xanthophores and melanophores (A and A'). When cells are transplanted from leotK3;spa;rse donors into pfe hosts, thereby creating chimeras with mutant xanthophores (present in the anterior part of the fish, left to the dotted line in B') and wild-type iridophores and melanophores, there is no wild-type pattern formed, but the chromatophores are almost uniformly distributed (B and B′) (9 clones in 72 fish). When cells are transplanted from leotK3;pfe;rse donors into nac hosts, thereby creating chimeras with mutant melanophores and wild-type iridophores and xanthophores (3 clones in 68 fish), again, no wild-type pattern is formed, but the mutant melanophores form loosely arranged fields of cells (C and C'). |