- Title
-
Basal keratinocytes contribute to all strata of the adult zebrafish epidermis
- Authors
- Lee, R.T., Asharani, P.V., and Carney, T.J.
- Source
- Full text @ PLoS One
|
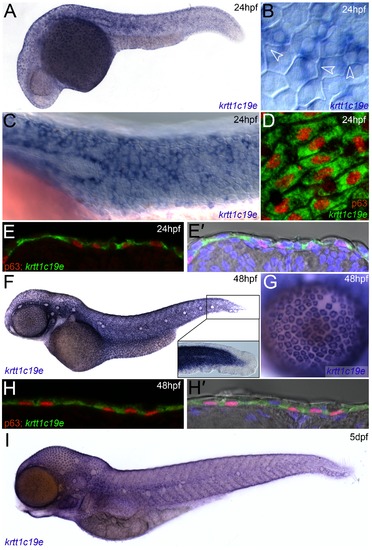
Expression of krtt1c19e in basal keratinocytes. In situ hybridisation of krtt1c19e at 24 hpf (A-E′), 48 hpf (F-H′) and 5 dpf (I), imaged laterally (A-D, F-G, I) or after cryosectioning (E-E′, H-H′). Overviews of embryos are shown at 24 hpf (A), 48 hpf (F) and 5 dpf (I), showing broad skin expression. Epidermal cells were visualised by counterstaining with DAPI (blue - E′, H′) or by Nomarski optics (B-C, F inset, E′, H′), and basal cell nuclei were immunolabelled using an antibody against ΔNp63 (red D-E′, H-H′). Strong krtt1c19e epidermal expression can be seen in the basal keratinocytes with the borders of overlying EVL cells intersecting basal cells (B - arrowheads). A gap in the krtt1c19e in situ signal is seen in the epidermis corresponding to the location of the migrating lateral line primordial (C). krtt1c19e expressing keratinocytes have ΔNp63 immunoreactive nuclei (D-E′, H-H′) and are seen below EVL cells in cryosections (E′, H′). A higher magnification of the tail region of the 48 hpf embryo is shown inset (F), with expression excluded from the fin epithelium, whilst a single layer of keratinocytes can be seen over the eye as part of the cornea (G). |
|
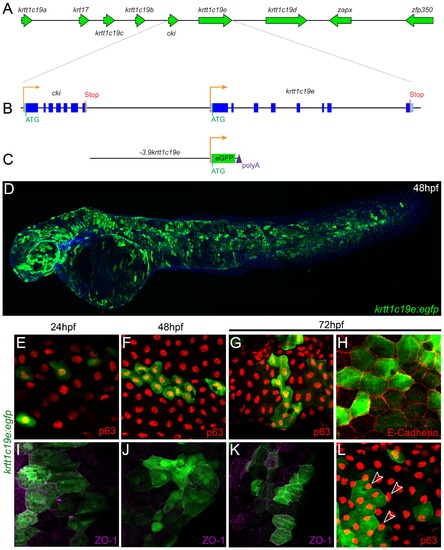
Isolation and transient activity of a krtt1c19e promoter. Map of the genomic context of the krtt1c19e gene in the type I keratin cluster on chromosome 19 (A) with a schematic of the intron/exon structure of the krtt1c19e gene and the upstream cki gene (B). The entire sequence upstream of krtt1c19e to the end of the cki was isolated and cloned upstream of egfp (C). Injection of this krtt1c19e:egfp construct into embryos yielded limited epidermal expression at 24 hpf but widespread eGFP expression by 48 hpf (C). E–L: Micrographs of eGFP positive epidermal cells in both the basal layer (E–H) and EVL (I–L), demonstrated by co-immunofluorescent labelling with antibodies against eGFP (green – E–L), ΔNp63 (red – E–G, L), E-cadherin (red - H) and ZO-1 (purple – I-K). At 24 hpf (E, I), 48 hpf (F, J) and 72 hpf (G–H, K–L) both ΔNp63 and E-cadherin positive basal cells are eGFP positive as are ZO-1 positive EVL cells. eGFP positive EVL cells can be seen above ΔNp63 positive nuclei (arrowheads - L). |
|
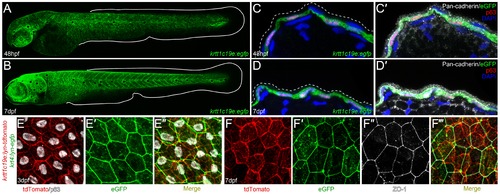
Characterization of krtt1c19e :egfp transgenic larvae. A-D′: Confocal images of eGFP expression in germline krtt1c19e:egfp transgenic larvae at 48 hpf (A, C-C′) and 7 dpf (B, D-D′). A-B: Lateral overviews of transgenic larvae indicating the krtt1c19e promoter drives eGFP expression in the epidermis of zebrafish larvae at all locations except the fins (extent of medial fins outlined by white line). C-D′: Immunofluorescent labelling of transgenic larvae cryosections demonstrates co-expression of eGFP (green; C-D′) and ΔNp63 (red; C-D′) in basal keratinocytes. Counterstaining with DAPI (blue; C-D′) and Pan-cadherin (white; C′, D′) highlights the eGFP negative region overlying EVL (demarcated by dashed lines; C′, D′). E-F′′′: Confocal images of the epidermis of germline krtt1c19e:lyn-tdtomato; krt4:lyn-egfp double transgenic larvae at 72 hpf (E-E′′) and 7 dpf (F-F′′′) immunofluorescently stained for eGFP (green; E′-E′′, F′, F′′′), tdTomato (red; E, E′′, F, F′′′), ΔNp63 (white; E, E′′) and ZO-1 (white; F′′-F′′′). The expression of membrane bound tdTomato delineates the ΔNp63 positive basal keratinocytes from the eGFP expressing ZO-1 positive EVL cells. |
|
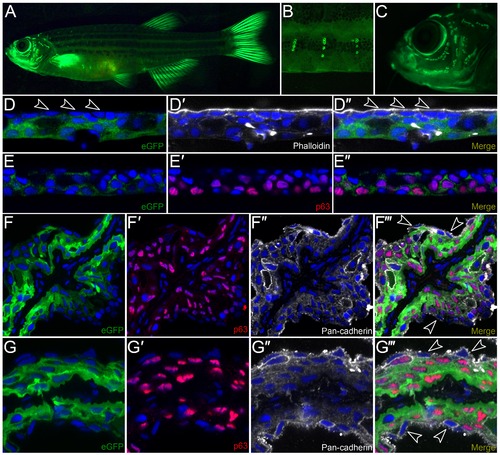
Characterization of krtt1c19e:egfp transgenic adult zebrafish. A-C: Lateral micrographs of krtt1c19e:egfp transgenic adult zebrafish, showing eGFP expression weakly in the trunk region but strongly in the fins (A). Expression is also seen associated with neuromasts of the lateral line system in the body (A, B) and head (C). D-G′′′: Immunostaining of cryosections from the trunk region (D-E′′) or fin (F-G′′′) of transgenic adults demonstrates expression is in basal and suprabasal epidermal cells, with no expression visible in the superficial epidermal stratum (arrowheads D-D′′). eGFP is shown in green (D, D′′, E, E′′, F, F′′′, G, G′′′) and co-localises with ΔNp63 (red; E′-E′′, F′, F′′′, G′, G′′′) in basal and suprabasal keratinocytes, and is excluded from the most superficial keratinocytes (e.g. arrowheads F′′′, G′′′) visualised by DAPI (blue; D-G′′′), Phalloidin staining (white, D′-D′′) or Pan-cadherin staining (white; F′′-F′′′, G′′-G′′′). EXPRESSION / LABELING:
|
|
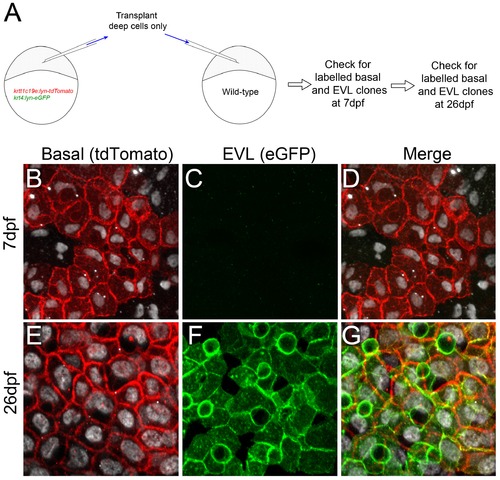
Transplantation of basal epidermal cells demonstrates contribution to the EVL at late stages. A: Schematic of transplantation strategy to test if the EVL of post metamorphosis larvae derives from the basal layer. Deep cells from krtt1c19e:lyn-tdtomato; krt4:lyn-egfp double transgenic embryos were transplanted into wild-type embryos after segregation of the EVL lineage at 4.3 hpf. Labelling of different epidermal cells was checked at 2 timepoints. B–G: Confocal images of the epidermis of representative recipient embryo at 7 dpf (B–D) and 26 dpf (E–G), immunostained for ΔNp63 (white; B,D–E, G), tdTomato (red; B,D–E, G) and eGFP (green; C–D, F–G). Transplanted deep cells only generate clones of basal cells expressing lyn-tdTomato at 7 dpf (B–D), but do not contribute to the EVL. The superficial layer is eGFP positive at 26 dpf indicating it is descended from the basal cells (E–G). n = 5 transplants were followed. |
|
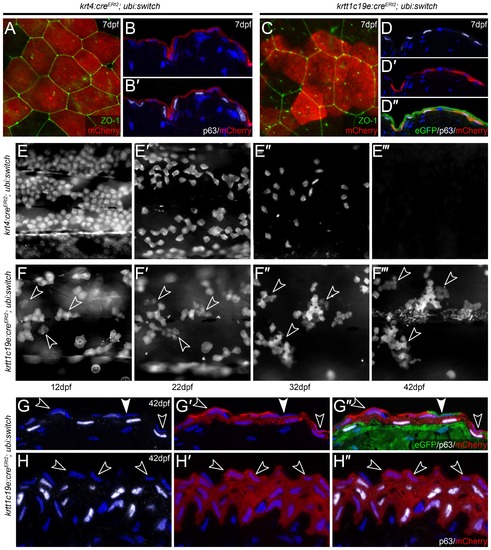
Embryonic EVL is lost and is replaced by cells from the basal epidermis during metamorphosis. A-D′′: Confocal images of lateral views (A, C) and transverse cryosections (B-B′, D-D′′) of krt4:CreERt2; ubi:swtch (A-B′) and krtt1c19e:CreERt2; ubi:swtch (C-D′′) at 5 dpf following 4-OHT mediated Cre conversion, and which have been immunofluorescently labelled with antibodies against mCherry (red; A-C, D′-D′′), ZO-1 (green; A, C), ”Np63 (white; B′, D, D′′) and eGFP (green; D′′) and counterstained with DAPI (blue; B-B′, D-D′′). Upon treatment of 4-OHT, krt4:CreERt2 drove recombination, and thus mCherry expression, in ZO-1-positive/”Np63-negative EVL cells (A-B′), whilst krtt1c19e:CreERt2 induced recombination in ΔNp63-positive/ZO-1-negative basal keratinocytes (C-D′′). E-F′′′: Time course of floxed krt4:CreERt2; ubi:swtch (E-E′′′) and krtt1c19e:CreERt2; ubi:swtch (F-F′′′) showing the same region of mCherry positive cells on the flank of representative individuals. Fluorescent images were taken at 12 dpf (E, F), 22 dpf (E′, F′) 32 dpf (E′′, F′′) and 42 dpf (E′′′, F′′′), and show that EVL cells are gradually lost, whilst clones of basal keratinocytes expand and stratify. n = 24 per genotype. G-H′′: Transverse cryosections of the trunk (G-G′′) and fin (H-H′′) epidermis of 42 hpf floxed krtt1c19e:CreERt2; ubi:switch transgenics, immunostained with antibodies against ΔNp63 (white; G, G′′, H, H′′), mCherry (red; G′-G′′, H′-H′′) and eGFP (green, G′′). Nuclei of all cells in the epidermis are marked by DAPI staining (blue; G-H′′). In contrast to 7 dpf (C-D′′), mCherry is now found in both suprabasal and the most superficial ΔNp63-negative cell layers (examples of the latter highlighted by open arrowheads). Occasional superficial cells, not derived from floxed basal cells, can be seen (green cell highlighted by closed arrowhead G-G′′). EXPRESSION / LABELING:
|
|
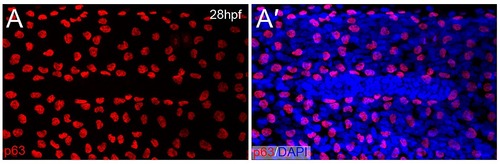
Lateral line primordium migration displaces basal epidermal cells. Confocal image of the lateral epidermis of a 28 hpf embryo immunofluorescently stained with an antibody against ΔNp63 (red; A-A′) and counterstained with DAPI (blue; A′). A gap in the basal epidermis is seen through displacement of the nuclei, and corresponds to the migrating primordium as indicated by the dense cluster of cell nuclei (A′). |
|
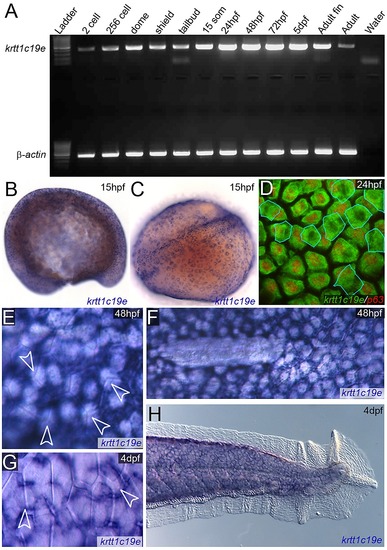
Timing of expression of krtt1c19e in the epidermis. A: RT-PCR of krtt1c19e (upper gel) at stages given and compared to β-actin positive control (lower gel) showing expression of krtt1c19e at all stages. Negative water control is given in far right lane. B–H: In situ hybridisations of krtt1c19e detected fluorescently (D) or by chromogenic precipitate (B–C, E–H) at 15 pf (B–C), 24 hpf (D), 48 hpf (E–F) and 4 dpf (G–H). Lateral (B) and dorsal (C) view of krtt1c19e in situ hybridisation at 15 hpf, when specific epidermal expression can be discerned. Counter-staining fluorescent in situ hybridisations with an antibody against ΔNp63 (red; D) demonstrates that in addition to the predominant basal keratinocyte expression, there is some low level expression of krtt1c19e in the EVL (outlined in blue) at 24 hpf. The predominant expression of krtt1c19e in basal layers at 48 hpf and 4 dpf is demonstrated through imaging the boundary of overlying EVL cells with Nomarski optics (arrowheads; E, G) and observing the lateral displacement of krtt1c19e expressing cells by the primordium at the end of its migration (F). The expression of krtt1c19e remains excluded from the epidermis of the medial fin at 4 dpf (H). |
|
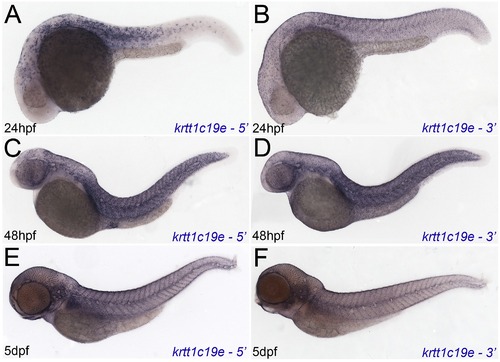
Independent in situ probes confirm krtt1c19e expression. Micrographs of 24hpf (A, B), 48hpf (C, D) and 5 dpf (E, F) embryos hybridised with 52 krtt1c19e (A, C, E) and 32 krtt1c19e (B, D, F) in situ probes. Whilst sensitivity was reduced, in particular at 24hpf, expression in the epidermis was identical to that seen with the full length probe. |
|
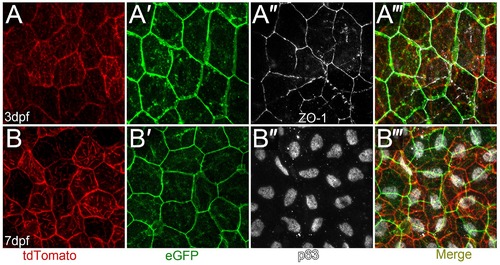
Mutually exclusive expression of lyn-tdTomato and lyn-eGFP in the basal layer and EVL in krtt1c19e:lyn-tdtomato; krt4:lyn-egfp double transgenics. A-B′′′: Confocal images of the epidermis of krtt1c19e:lyn-tdtomato; krt4:lyn-egfp double transgenic larvae at 3 dpf (A-A′′′) and 7 dpf (B-B′′′) immunofluorescently stained for eGFP (green; A′, A′′′, B′, B′′′), tdTomato (red; A, A′′′, B, F′′′), ZO-1 (white; A′′-A′′′) and ”Np63 (white; B′′, B′′′). The krtt1c19e promoter drives expression in the ”Np63 positive basal layer. |
|
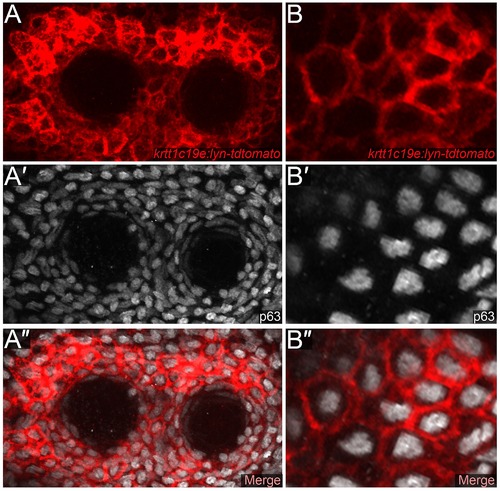
Strong expression of the krtt1c19e promoter in epidermal cells surrounding the adult neuromasts. Low (A-A′′) and high (B-B′′) magnification confocal images of cells surrounding trunk neuromasts of a krtt1c19e:lyn-tdtomato transgenic adult, immunofluorescently stained for tdTomato (red; A, B, A′′, B′′) and ΔNp63 (white; A′, B′, A′′, B′′). High level promoter activity is evident in p63 positive epidermal cells surrounding the neuromasts. |