- Title
-
Kif11 dependent cell cycle progression in radial glial cells is required for proper neurogenesis in the zebrafish neural tube
- Authors
- Johnson, K., Moriarty, C., Tania, N., Ortman, A., Dipietrantonio, K., Edens, B., Eisenman, J., Ok, D., Krikorian, S., Barragan, J., Golé, C., and Barresi, M.J.
- Source
- Full text @ Dev. Biol.
|
Characterization of kif11 expression in radial glia and its effect on radial glial soma localization. (A–D) kif11 siblings were grown to 30 hpf and labeled for radial glia (Gfap) and acetylated Tubulin (AT) to identify glial and axon defects. In wild type siblings a small population of Gfap+ radial glial somas can be visualized at the luminal surface in a maximum intensity projection of optical sections along the parasagittal plane that encompass the ventricular zone (A) as well as in cross section of the neural tube (B). An increase in the number of radial glial somas can be seen the neural tube of kif11-/- mutants (C,D). (E) Quantification of Gfap+ somas counted from maximum intensity projections of parasagittal optical slices within the neural tube and the total number of nuclei in transverse cross sections of the neural tube showed an 8-fold increase in the number of Gfap+ cell bodies in kif11-/- mutants, but no change in the number of total nuclei between siblings. (F) RT-PCR analysis of kif11 expression between pooled wild type/heterozygote siblings and kif11-/- at 48 hpf. β-actin was used as a loading control. (G–M) Wild type kif11 mRNA was present throughout the forebrain (G, lateral; H, dorsal) and trunk (I, J–M) at 27 hpf, concentrated at the luminal surface of the VZ (J,K) and into the floor plate (J,K, insets). (K–M) Fluorescent kif11 detection (K) within a Tg(gfap:EGFP) embryo (L) shows that kif11 mRNA is present down the length of the ventricular zone and colocalizes with cells expressing GFP associated with the lumen of the VZ (M). |
|
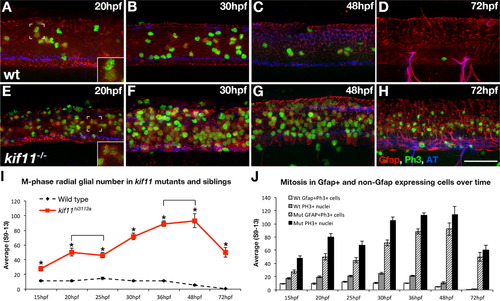
Loss of kif11 causes an accumulation of radial glia in mitosis. (A–H) Lateral views of kif11-/- sibling neural tubes fixed at timepoints throughout neurogenesis and labeled for radial glia (Gfap), Phosphohistone H3 (PH3) and acetylated Tubulin (AT). Within all siblings, most Gfap+ cells labeled with our antibody were PH3+, indicating these cells were actively proliferating at the time of fixation (A,E insets). At all time points assayed, there were significantly more Gfap+/PH3+ cells in kif11-/- embryos compared with wild type siblings (E–H, quantified in I, asterisks), as well as more PH3+ nuclei (J, shaded and black bars). No change in the number of Gfap+ cells was seen between 20–25 hpf and 36–48 hpf within kif11-/- (I, brackets). Of all proliferating cells (J), it was noted that an unidentified population (PH3+, Gfap) is also kif11 dependent in both wild type and kif11-/- neural tubes. Error bars delineate the standard error of the mean. Asterisks indicate a statistical significance (t-test, two tailed, assuming equal variances, p<0.05). EXPRESSION / LABELING:
|
|
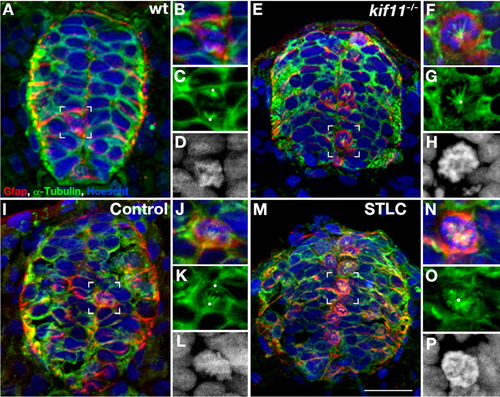
Loss of Kif11 causes monoaster spindle formation in radial glial cells during division. kif11 siblings and wild type embryos treated with either STLC or DMSO control from 5 to 30 hpf were labeled for radial glia (Gfap), microtubules (α-Tubulin) (microtubules), and histones (Hoechst stain) to identify mitotic spindle phenotypes. (A) kif11 wild type siblings and (I) vehicle control (DMSO) treated embryos exhibit normal spindles in M-phase (enlarged in B–D and J–L, respectively) while (E) kif11-/- mutants and (M) STLC treated embryos display monoastral spindles characteristic of mitotic arrest (enlarged in F–H and N–P, respectively). White dots designate presumptive locations of centrosomes. For greater clarity Hoechst is displayed as a greyscale image in the enlargements (D, H, L, P). |
|
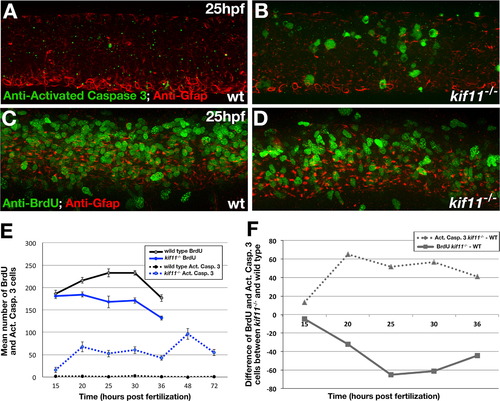
kif11 mutants have inversely proportional changes to the number of cells dying and proliferating. (A,B) Lateral view of the 25 hpf neural tube labeled for anti-activated Caspase-3 (green) and anti-Gfap (red) antibodies in wild type (A) and kif11-/- mutants (B). (C,D) Lateral view of the 25 hpf neural tube labeled for anti-BrdU (green) and anti-Gfap (red) antibodies in wild type (C) and kif11-/- (D). (E) Quantification of cell death (dashed lines) and BrdU incorporation (solid lines). Anti-activated Caspase-3 labeling was increased in kif11-/- mutants from 15 hpf to 72 hpf (E, dashed blue line), whereas the number of cells in S-phase was reduced from 20 hpf to 36 hpf in kif11-/- mutants (E, solid black line) as compared with wild type embryos (black lines). (F) Graphical representation of the difference between the number of cells dying (dashed line) or BrdU positive (solid line) between kif11 siblings. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Kif11 function is most required during two distinct periods for proper cell division. (A) Schematic illustrating time of exposure to S-trityl-⌊-cysteine (STLC) or vehicle control (dimethyl sulfoxide) and duration of treatment between groups. (B–G) Lateral views of post-treatment embryos labeled for radial glia (Gfap), Ph3 and AT. (H) In all test groups, there were significantly more Gfap+/Ph3+ and Gfap-/Ph3+ cells in STLC treated embryos (E–G) compared with their controls (B–D) at 5–30 hpf, 10–30 hpf, 15–30 hpf, 20–30 hpf, and 25–30 hpf. Error bars delineate the standard error of the mean. Asterisks indicate a statistical significance (t-test, two tailed, assuming equal variances, p<0.05). EXPRESSION / LABELING:
|
|
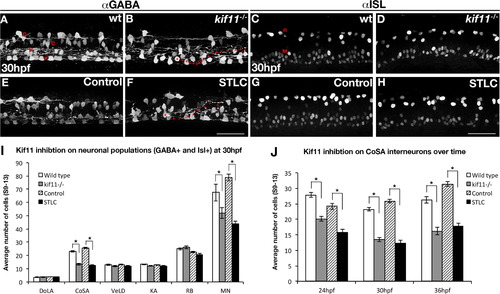
Kif11 is required for motorneurons and CoSA interneurons. Wild type/vehicle control (A,C,E,G) and kif11-/-/STLC treated embryos (B,D,F,H) were labeled for anti-GABA (A,B,E,F) to label distinct DoLA, CoSA, VeLD, and KA interneurons (denoted D,C,V, and K, respectively), as well as anti-ISL (C,D,G,H) to label Rohon Beard sensory neurons and motor neurons (denoted R and M, respectively). Decreases in GABA+ cell populations as well as axon pathfinding errors were found in Kif11 deficient embryos as compared with controls (asterisks in B and F label cell body, arrowheads mark axon trajectory). (I) Quantification of neuronal populations identified decreases specifically in CoSA and MN populations within Kif11 deficient embryos (asterisks, p<0.05) as compared with controls. (J) Time course analysis of GABA+ cell types at 24 hpf, 30 hpf and 36 hpf shows a consistent number of GABA+ cells both lost and retained over time in kif11 mutants and STLC treated embryos. |
|
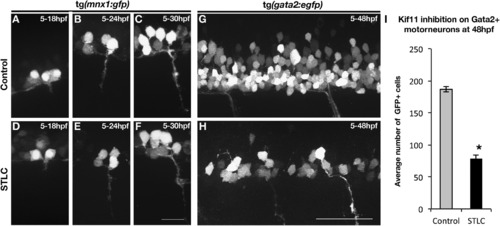
Kif11 is required for secondary motorneurons but not primary motorneurons. (A–F) Visualization of primary motorneuron development in somite 11 over time using the Tg(mnx1:GFP) transgenic line. No difference in the formation of primary motorneurons was observed in STLC treated Tg(mnx1:GFP) transgenic embryos (D–F) as compared to control DMSO treated embryos (A–C). (G,H) Lateral view of the neural tube of Tg(gata2:EGFP) transgenic embryos at 48 hpf to assess secondary motorneurons in control DMSO treated (G) or STLC treated transgenic embryos from 5 to 48 hpf (H). (I) A significant decrease in GFP+ neurons was seen in STLC treated Tg(gata2:EGFP) embryos. Error bars delineate the standard error of the mean. Asterisks indicate a statistical significance (t-test, two tailed, assuming equal variances, p<0.05). EXPRESSION / LABELING:
PHENOTYPE:
|
|
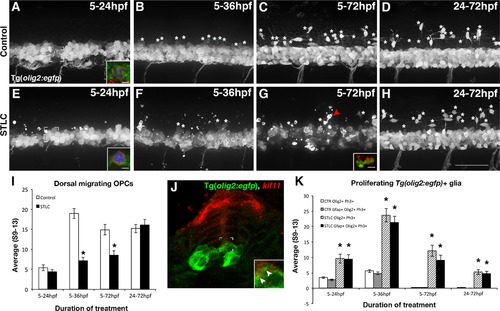
Kif11 is required for oligodendrocyte development. Lateral views of Tg(olig2:EGFP) embryos treated with either vehicle control (A–D) or STLC (E–H) at 5hpf and fixed at time points critical for oligodendrocyte development (5–24 hpf, 5–36 hpf, 5–72 hpf, and 24–72 hpf). Dorsal migrating OPCs (white asterisks) were present in control embryos older than 24 hpf (B–D), which were significantly reduced in STLC treated groups (F–H, quantified in I, black asterisks p>0.05), except when treated from 24 to 72 hpf (no statistical significance from the vehicle control, p>0.51). Irregular GFP+ fragments (G arrowhead) co-labeled with anti-Activated Caspase-3 (G, inset, red fluorescence) indicative of cell death. (J) kif11 mRNA detection within 27 hpf Tg(olig2:EGFP) embryos indicated that a small portion of olig2 cells associated with the ventricular zone colocalize with kif11 expression (J inset, arrowheads). Triple-labeled (GFP+, Ph3+, Gfap+) cells were identified in most control and treatment groups (A,E brackets; insets are maximum intensity projection of extracted Z-stack series in A,E; quantified in K), and were significantly increased in all STLC treated groups (K, asterisks). Error bars delineate the standard error of the mean. Asterisks indicate a statistical significance (t-test, two tailed, assuming equal variances, p<0.05). EXPRESSION / LABELING:
|
|
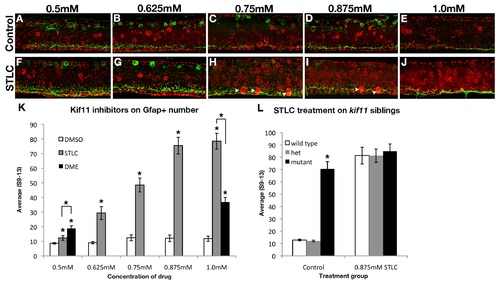
STLC treatment phenocopies kif11-/- mutants without causing additional morphological deformations. Lateral views of wild type neural tubes after treatment from 5hpf to 30hpf with various concentrations of either the vehicle control (dimethyl sulfoxide, A-E) or S-trityl-L-cysteine (STLC, F-J). Astroglial and axon changes were visualized by labeling for Gfap and acetylated Tubulin (AT). No change in radial glial soma number was seen in DMSO control embryos at any concentration tested. Gfap+ floorplate somas (arrowheads) were only seen at higher concentrations of STLC (H-J). (K) Quantification of the average Gfap+ soma number following Kif11 inhibition. The lowest concentration of DME (0.5mM) caused an increase in GFAP+ cell bodies, however extreme gross morphological defects were also noted therefore this drug was not further processed. Gfap+ cell body number was increased in STLC concentrations higher than 0.625mM (grey bars, asterisks). (L) A maximum concentration of 0.875mM phenocopied the kif11 mutant. No additive effects were seen or quantified when kif11-/- mutants were treated with 0.875mM STLC (L, black bars, data not shown). There was no statistically significant difference seen between STLC-treated wild type, heterozygous kif11+/-, and homozygous kif11-/- (L). Error bars delineate the standard error of the mean. Asterisks indicate a statistical significance (t-test, two tailed, assuming equal variances, p<0.05). |
|
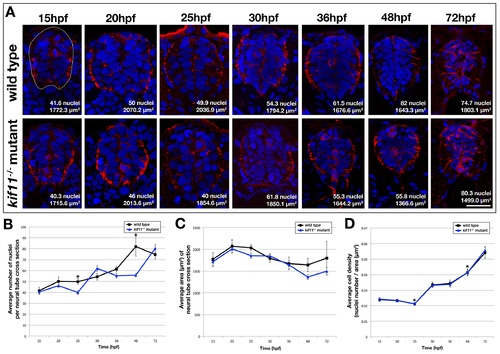
Cell density in the kif11-/- neural tube is unchanged. (A) Cross sectional analysis of Gfap-positive astroglia (red) and Hoechst-positive nuclei (blue) in the developing spinal cord over time from 15hpf to 72hpf in wild type (top row) and kif11-/- mutants (bottom row). The cross sectional area of the neural tube was calculated for each section following manual outlining of the neural tube, which was based on Gfap labeling that delineates the outer edges of the neural tube (white line shown in 15hpf wild type image). (B-D) Quantification and data analysis of cell densities in the neural tube. Three sections corresponding to the axial locations of approximately somite 4-6, 10-12, and 16-18 were collected per embryo. 4 embryos were counted per time point. A resulting total of 12 sections were counted for 15-30hpf and 48hpf time points, and 11 sections were counted for 36hpf and 10 sections for 72hpf. Nuclei and area counts were averaged for each time point, and are shown on each example image (A). Only the 25hpf and 48hpf time points showed a statistically significant decrease in the number of nuclei in the kif11-/- mutant (blue line) neural tube as compared with wild type controls (black line) (B, asterisks). No statistically significant difference was seen in the cross sectional area of the neural tube between kif11 siblings (C). (D) Calculated cell density in the neural tube showed no visible difference between kif11 siblings (C, overlapping blue and black lines); however both 25hpf and 48hpf continued to be statistically significantly different (asterisks; 25hpf, p=0.004; 48hpf, p=0.004). Error bars delineate the standard error of the mean. Asterisks indicate a statistical significance (t-test, two tailed, assuming equal variances, p<0.05). |
Reprinted from Developmental Biology, 387(1), Johnson, K., Moriarty, C., Tania, N., Ortman, A., Dipietrantonio, K., Edens, B., Eisenman, J., Ok, D., Krikorian, S., Barragan, J., Golé, C., and Barresi, M.J., Kif11 dependent cell cycle progression in radial glial cells is required for proper neurogenesis in the zebrafish neural tube, 73-92, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.