- Title
-
Par3 controls neural crest migration by promoting microtubule catastrophe during contact inhibition of locomotion
- Authors
- Moore, R., Theveneau, E., Pozzi, S., Alexandre, P., Richardson, J., Merks, A., Parsons, M., Kashef, J., Linker, C., and Mayor, R.
- Source
- Full text @ Development
|
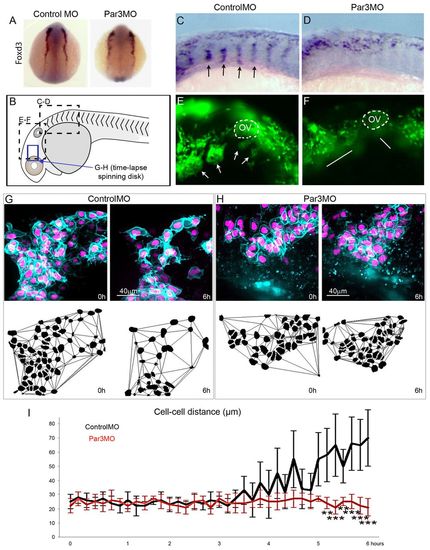
Par3 is required for NC migration in zebrafish. (A) Dorsal view of 5-somite stage zebrafish embryos injected unilaterally with ControlMO or Par3MO and processed for in situ hybridisation against foxd3; no effect on NC induction was observed (n=72). (B) Schematic representation of zebrafish embryo indicating the regions shown in C-H. (C,D) Lateral views showing foxd3 expression in the trunk of ControlMO-injected (C) or Par3MO-injected (D) 20-somite embryos. Arrows indicate distinct trunk NC streams. (E,F) Lateral views showing heads of 24-hour sox10:egfp embryos injected with ControlMO (E) or Par3MO (F). OV, otic vesicle. Arrows indicate distinct cranial NC streams. Lines indicate that no distinct streams are observed. (G,H) Single frames from time-lapse movies showing one cranial NC stream and corresponding Delaunay triangulation at 0 hours (15 somites) and 6 hours from ControlMO-injected (G) and Par3MO-injected (H) embryos. (I) Distance between cranial NC cells analysed by nearest neighbour computation. n=15 embryos for each condition, with more than 30 cells analysed per explant; **P<0.01, ***P<0.001. Error bars indicate s.d. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Par3 inhibition does not affect cell adhesion in Xenopus or zebrafish. (A-F) Cell adhesion molecules analysed in Xenopus embryos. (A-C) Immunostaining against β-catenin in control (A) or Par3MO-injected NC cells (B). (C) Pixel intensity of β-catenin immunostaining was measured across the contact and normalised to the average value background levels 5 μm away from the contact for each image. There is no difference in pixel intensity between control cells (n=84 contacts) and Par3MO-injected cells (n=44 contacts; P>0.05 at all distances from contact). (D-F) Control (D) or Par3MO-injected (E) NC cells expressing p120-cateninGFP. (F) Pixel intensity analysis as described in C. There is no difference in pixel intensity of p120-catenin immunostaining between control cells (n=24 contacts) and Par3MO-injected cells (n=41 contacts; P>0.05 at all distances from contact). (G-L) N-cadherin immunostaining in control (G-I) or Par3MO injected (J-L) zebrafish embryos. sox10GFP transgenic embryos were used to identify NC cells. Note that N-cadherin staining is not affected by Par3MO in the NC. n=150 embryos, fixed at 24 hpf. (M-P) Cell adhesion is not affected by Par3MO. (M) Schematic representation of the cell reaggregation assay. (N) Control Xenopus NC cells. (O) Control/Par3MO Xenopus NC cells. (P) Control/N-cadherinMO Xenopus NC cells. Scale bars: 10 μm. EXPRESSION / LABELING:
|
|
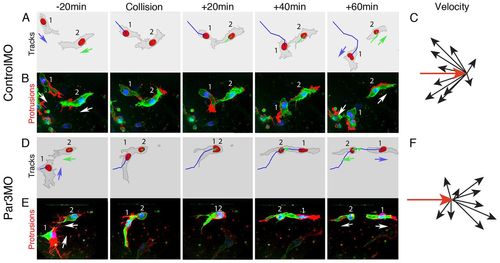
Par3 is required for CIL in vivo in zebrafish. Single frames from time-lapse movies illustrating a collision between two cranial NC cells (1 and 2) in a zebrafish embryo injected with ControlMO (A,B) or Par3MO (D,E). Images are overlapped with tracks of migratory paths (A,D) or the difference in protrusion between current and previous frames (B,E). Red area indicates new protrusions. (C,F) Velocity vectors of colliding NC cells in ControlMO-injected (C) or Par3MO-injected (F) embryos. Cell position was recorded at 5-minute intervals. Velocity change was clustered in ControlMO-injected cells (n=10; P<0.05), but no difference in the velocity change was observed in Par3MO-injected cells (n=9; P>0.1). Red arrow indicates the initial velocity vector. EXPRESSION / LABELING:
PHENOTYPE:
|