- Title
-
Lypd6 Enhances Wnt/beta-Catenin Signaling by Promoting Lrp6 Phosphorylation in Raft Plasma Membrane Domains
- Authors
- Özhan, G., Sezgin, E., Wehner, D., Pfister, A.S., Kühl, S.J., Kagermeier-Schenk, B., Kühl, M., Schwille, P., and Weidinger, G.
- Source
- Full text @ Dev. Cell
|
Zebrafish lypd6 Is a Wnt/β-Catenin Target Gene that Codes for a GPI-Anchored Plasma Membrane Protein (A) lypd6 whole-mount in situ hybridization (WMISH) showing downregulation in transgenic embryos expressing Axin1 or Dkk1 at gastrula and somitogenesis stages (n [85% epiboly]: Axin1 16/17 embryos, Dkk1 19/19; n [10-somites]: Axin1 21/21, Dkk1 18/18). hs, heat shock. (B) lypd6 expression levels determined by qRT-PCR in hs:Axin1, hs:Dkk1, and hs:Wnt8 transgenic embryos treated as in (A) and shown relative to those in wild-type embryos. Error bars, SEM. (C) Domain structure of N-terminally GFP-tagged wild-type (spGFP-Lypd6) and C-terminally truncated Lypd6 (spGFP-Lypd6ΔGPI). (D) Localization of spGFP-Lypd6 and spGFP-Lypd6ΔGPI in enveloping layer (EVL) cells of dome stage zebrafish embryos and in Chinese hamster ovary (CHO) cells. (E) Release of spGFP-Lypd6 and spGFP-GPI from the plasma membrane of HEK293T cells upon treatment with phosphatidylinositol-specific phospholipase C (PiplC). spGFP-Lypd6ΔGPI and spGFP are detectable in the conditioned media without PiplC treatment. See also Figure S1. EXPRESSION / LABELING:
|
|
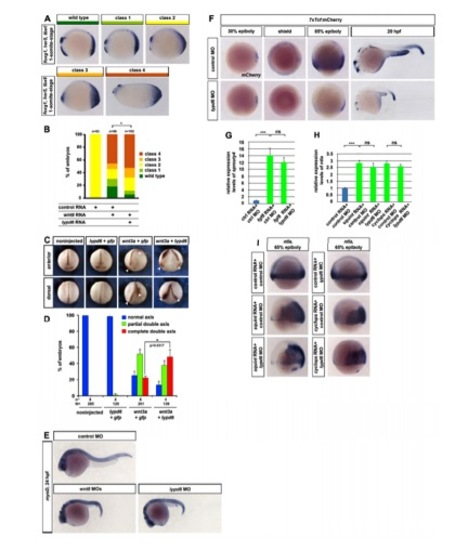
lypd6 Enhances Wnt/β-Catenin Signaling (A) Classification of phenotypes caused by wnt8 overexpression in zebrafish embryos: class 1, small eyes (arrow); class 2, no eyes (arrow) but normal midbrain-hindbrain boundary (mhb) (arrowhead); class 3, severely reduced forebrain and midbrain and mhb not detectable (arrowhead); and class 4, loss of notochord. (B) Phenotypes in hs:Wnt8 transgenic embryos injected with spGFP-lypd6 RNA (150 pg), spGFP-lypd6ΔGPI (140 pg), or equimolar amounts of spGFP-GPI control RNA and heat shocked at shield stage and scored at 24 hpf. (C) pBAR luciferase reporter activity normalized to renilla luciferase levels in Wnt3a CM-treated HEK293T cells transfected with spGFP-Lypd6 (100 ng), spGFP-Lypd6ΔGPI (95 ng), or equimolar amounts of spGFP-GPI control. Error bars, SEM. (D) Morphological phenotypes at 24 hpf of embryos injected with lypd6 MO (10 ng, 42/51 embryos) are rescued by coinjected lypd6 RNA (150 pg, 49/58 embryos). (E) Rescue of phenotypes in hs:Wnt8 embryos injected with lypd6 MO (10 ng). Coinjected lypd6 RNA (150 pg) restores the more severe phenotypes. (F) Reduction of reporter activity in 7xTcf:mCherry embryos injected with 10 ng lypd6 MO plus 100 ng control RNA (n = 28/31). Coinjected lypd6 RNA (75 ng) restores expression (n = 21/25). (G) Downregulation of cdx4 (90% epiboly, 19/20) and sp5l (80% epiboly, 13/15) in embryos injected with 10 ng lypd6 MO. See also Figure S2. PHENOTYPE:
|
|
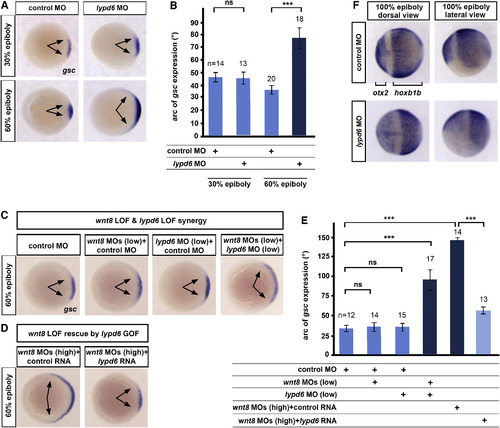
lypd6 Is Necessary for wnt8-Mediated Mesoderm and Neuroectoderm Patterning (A) Expansion of the dorsal organizer domain (arrows) marked by goosecoid (gsc), not at late blastula (30% epiboly), but at early gastrula (60% epiboly) stage in embryos injected with 10 ng lypd6 MO. (B) Quantification of the experiment shown in (A). Error bars, SEM. (C) Changes in the size of the gsc expression domain (arrows) at 60% epiboly in embryos injected with low doses of wnt8.1 and wnt8.2 MOs (1.25 ng each) and lypd6 MO (2 ng), relative to embryos injected with equimolar amounts of control MO. (D) gsc expression in embryos injected with high doses of wnt8 MOs (5 ng each) and 150 pg lypd6 RNA or equimolar amounts of GFP control RNA. (E) Quantification of the experiments shown in (C) and (D). Error bars, SEM. (F) Expansion of the forebrain marker otx2 and reduction of the posterior neural marker hoxb1b at the 100% epiboly stage in embryos injected with 10 ng lypd6 MO (19/23). EXPRESSION / LABELING:
|
|
lypd6 Acts in Wnt-Receiving Cells at the Level of the Wnt Receptor Complex (A) pBAR luciferase reporter activity in mouse L cells treated with CM produced from control or Wnt3a-producing L cells that were transfected with lypd6 or control siRNA. Error bars, SEM. (B) pBAR luciferase reporter activity in Wnt3a CM treated L cells transfected with lypd6 siRNA. Error bars, SEM. (C) lypd6 enhances Wnt/β-catenin signaling upstream of the β-catenin degradation complex. Coinjection of lypd6 RNA rescues the reduction in reporter activity in 7xTcf:mCherry embryos caused by wnt8 MOs (5 ng each, 17/22), but not by RNAs of axin1 (50 pg, 19/19), gsk3β (150 pg, 17/20), or dominant negative tcf3, tcf3ΔC (125 pg, 23/24) at the 85% epiboly stage. See also Figure S3. |
|
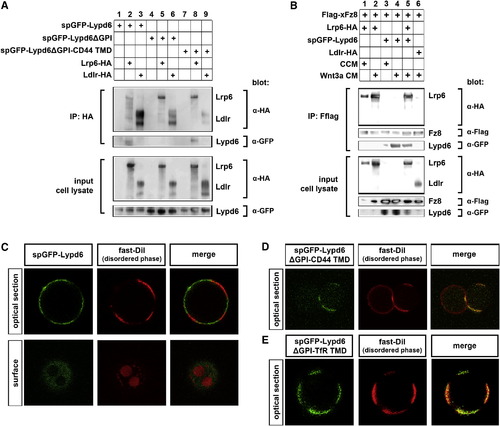
Lypd6 Interacts with Lrp6 and Partitions into Ordered Membrane Domains in Giant Plasma Membrane Vesicles (A) spGFP-Lypd6 and spGFP-Lypd6ΔGPI-CD44 TMD coIP with Lrp6-HA, but not with Ldlr-HA, in HEK293T cells. spGFP-Lypd6ΔGPI does not coIP with Lrp6-HA or Ldlr-HA. (B) spGFP-Lypd6 coIPs with Flag-xFz8 in HEK293T cells, and this binding is strongly enhanced by Wnt3a. Flag-xFz8 also binds to Lrp6-HA, which is enhanced by Wnt3a, but does not bind to Ldlr-HA. Lypd6 and Lrp6 still bind to Fz8 when expressed together. (C–E) Partitioning of various GFP-tagged Lypd6 constructs in giant plasma membrane vesicles (GPMVs) derived from CHO cells. Fast DiI marks the disordered compartments. See also Figure S4. |
|
Lypd6 Knockdown or Mislocalization to Nonraft Membrane Domains Shifts Lrp6 Phosphorylation to These Domains and Inhibits Wnt Signaling (A) pBAR activity in HEK293T cells transfected with Wnt8 (20 ng) plus spGFP-Lypd6 (100 ng) or spGFP-Lypd6ΔGPI-CD44 TMD (95 ng) or equimolar amounts of spGFP-GPI control. Error bars, SEM. (B) Classes of phenotypes in wnt8-overexpressing embryos injected with wnt8 (20 pg) plus spGFP-lypd6 (150 pg) or spGFP-lypd6ΔGPI-TfR TMD (140 pg) or equimolar amounts of spGFP-GPI control RNA. spGFP-lypd6ΔGPI-TfR TMD significantly rescues Wnt8-induced phenotypes. Class 5, hyperdorsalization. (C) spGFP-Lypd6ΔGPI-CD44 TMD reduces Wnt3a-induced phosphorylation of Lrp6 at S1490 in HEK293T cells assayed at 6 hr poststimulation with Wnt3a CM. (D) lypd6 esiRNA reduces Wnt3a-induced Lrp6 phosphorylation in HEK293T cells. (E) Fractionation of plasma membrane derived from HEK293T cells treated with Wnt3a into soluble and detergent-resistant fractions (DRMs). Endogenous TfR2 (a marker for soluble fractions), Caveolin-1 (Cav1, marking DRMs), P-Lrp6 (S1490), Lrp6, and overexpressed GFP in spGFP-Lypd6 (5 μg) or spGFP-Lypd6ΔGPI-TfR TMD (4.6 μg) or control spGFP-GPI (2.9 μg) transfected cells were detected by western blotting. (F) Fractionation of plasma membrane derived from HEK293T cells transfected with 2 μg EGFP control or lypd6 esiRNAs and treated with Wnt3a CM into soluble and DRM phases. (G) Imaging and quantification of Wnt3a- or Dkk1-induced Lrp6 speckles in HEK293T cells stably expressing Lrp6-EGFP. Scale bar, 10 μm. See also Figure S5. |
|
lypd6 expression during zebrafish and Xenopus embryonic development and bioinformatic analyses of Lypd6 protein sequence. (A-D) Zebrafish lypd6 expression pattern detected by whole mount in situ hybridization (WMISH) at the indicated stages. ov = otic vesicle, opv = optic vesicle, hpf = hours post fertilization. EXPRESSION / LABELING:
|
|
Lypd6 specifically enhances Wnt/β-catenin signaling in zebrafish and Xenopus embryos. (A-B) wnt8-induced anteroposterior patterning defects are enhanced by lypd6 overexpression during zebrafish gastrulation. |
|
Lypd6 is required for Wnt/β-catenin signaling in Wnt-receiving cells. (A) Mouse lypd6 transcripts are knocked down to about 27% in murine L cells transfected with lypd6 siRNA as determined by qRT-PCR at 48 hours after transfection. |
|
Lypd6 directly interacts with Lrp6, while it binds to Fz8 in a Wnt3a-dependent manner but does not interact with Wnt8. (A) Purified secreted Lypd6 (spGFP-Lypd6ΔGPI) co-IPs with secreted recombinant hLrp6-Fc chimeric protein (lane 2), while secreted GFP does not (lane 1). Dkk1-Flag can also be co-IPed with hLrp6-Fc (lane 3). |
Reprinted from Developmental Cell, 26(4), Özhan, G., Sezgin, E., Wehner, D., Pfister, A.S., Kühl, S.J., Kagermeier-Schenk, B., Kühl, M., Schwille, P., and Weidinger, G., Lypd6 Enhances Wnt/beta-Catenin Signaling by Promoting Lrp6 Phosphorylation in Raft Plasma Membrane Domains, 331-345, Copyright (2013) with permission from Elsevier. Full text @ Dev. Cell