- Title
-
Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration
- Authors
- Fang, Y., Gupta, V., Karra, R., Holdway, J.E., Kikuchi, K., and Poss, K.D.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
Translational profiling of cardiomyocytes during zebrafish heart regeneration. (A) Tissue section indicating localization of EGFP-L10a fluorescence to cardiomyocytes in cmlc2:TRAP hearts. (B) Higher-magnification view of an uninjured cmlc2:TRAP ventricular apex. (C) Regeneration appears normal in cmlc2:TRAP ventricles 30 d after partial resection. Dashed line indicates approximate amputation plane. (Scale bars for A–C, 50 μm.) (D) RT-PCR of total or immunoprecipitated (TRAP) RNAs isolated from adult cmlc2:TRAP ventricles. β-actin1, hand2, cmlc2, ventricular myosin heavy chain (vmhc) and anf (also known as natriuretic peptide precursor A) are expressed in cardiomyocytes (CMs), and flk1 (endocardial), gata2a (hematopoietic), and tcf21 (epicardial) are noncardiomyocyte genes (Non-CMs). (E) Heat map from microarray indicating increased levels of Jak1/Stat3 pathway members in cmlc2:TRAP RNA samples at 1 dpa. (F) qPCR using RNAs immunoprecipitated from cmlc2:TRAP ventricles, confirming up-regulation of Jak1/Stat3 pathway members after injury. Expression levels were normalized to that of β-actin2, and further normalized to that of the uninjured sample. Data are mean ± SEM n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, Student t test (unpaired, two-tailed). (G) Western Blots using total proteins extracted from uninjured and 1-dpa hearts, indicating increased protein levels of Stat3, phosphorylated Stat3 (P-Stat3), and Bcl2 at 1 dpa. Tubulin was used as a loading control (shown for Stat3 and P-Stat3 lanes). EXPRESSION / LABELING:
|
|
Induction of Jak1/Stat3 pathway members and ligands after cardiac injury. (A and B) In situ hybridization revealing Jak1/Stat3 pathway members (shown here are il6st and socs3b) induced in an organ-wide manner at 1 dpa (A), and then restricted to the injury site at 7 dpa (B). Dashed lines indicate approximate amputation plane. Brackets indicate injury site. (C) qPCR from ventricular RNA samples revealed induction of il11a, il11b, and lif at 1 and 7 dpa. Data are mean ± SEM n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, Student t test (unpaired, two-tailed). Expression levels were normalized to that of β-actin2, and further normalized to that of the uninjured sample. (D) il11a expression was induced in an organ-wide manner in endocardial cells at 1 dpa, and localized to the injury site at 7 dpa. Dashed line indicates approximate amputation plane. Brackets indicate injury site. (E) RT-PCR of samples from purified endocardial/endothelial (fli1:EGFP+) cells or pu.1-enriched cells at 1 dpa. il11a was detected in fli1:EGFP+ samples, similar to the Vegf receptor flk1. lif was detected in pu.1-enriched samples; it was undetectable when the same amount of total ventricular RNA was used for amplification (total). il11b was detected in pu.1-enriched samples and less so in fli1:EGFP+ samples. anf and tcf21 are markers for cardiomyocytes and epicardium, respectively, and were detected in total ventricular RNA samples. (Scale bars, 50 μm.) EXPRESSION / LABELING:
|
|
Cardiomyocyte Stat3 activity is essential for heart regeneration. (A) Cartoon representation of transgenes used for inducible expression of a dominant-negative Stat3 (dnStat3-GFP) cassette in cardiomyocytes. (B) cmlc2:CreER;β-act2:RSdS (dnStat3) and β-act2:RSdS (control) clutchmates were administered 4-HT, injured 3 d later, and collected for histological analysis of muscularization and scarring at 30 dpa. Muscle regeneration was blocked, and wounds healed by fibrin retention and scar formation in dnStat3-expressing fish (n = 9). MHC, myosin heavy chain. Dashed lines indicate approximate amputation plane. (C) Confocal images of sections from 7dpa ventricles, stained with antibodies against PCNA (a proliferation marker) and Mef2 (a cardiomyocyte marker). (Insets) Higher-magnification images of the squares; arrows, proliferating cardiomyocytes. (D) Quantification of cardiomyocyte (CM) proliferation during regeneration (7 dpa) or after 10 d of rapid or slow (normal) growth conditions. Ventricular resection in adults triggered similar levels of cardiomyocyte proliferation as rapid growth conditions in juveniles/young adults. However, inhibitory effects of dnStat3 expression were apparent only during injury-induced regeneration. Data are mean ± SEM n = 6 (regeneration) or 12 (growth), *P < 0.05, ***P < 0.001. Student t test (unpaired, two-tailed). (Scale bars, 50 μm.) |
|
Evidence that Rln3a acts downstream of Stat3 during heart regeneration. (A) qPCR using cmlc2:TRAP RNA samples, indicating that rln3a is highly induced in cardiomyocytes (CMs) at 1 dpa. Data are mean ± SEM n = 3, **P < 0.01, Student t test (unpaired, two-tailed). Expression levels were normalized to that of β-actin2, and further normalized to that of the uninjured sample. (B) qPCR indicating reduced rln3a levels in dnStat3-expressing fish at 1 and 7 dpa. Data are mean ± SEM n = 3, ***P < 0.001, Student t test (unpaired, two-tailed). Expression level was normalized to that of β-actin2, and further normalized to that of the uninjured control sample. (C and D) Enrichment of Stat3 at its predicted binding sites in the rln3a promoter in 1-dpa ventricular tissue. ChIP was performed using anti-Stat3 or IgG antibodies. Immunoprecipitated genomic DNA was analyzed by PCR (C) and qPCR (D). Genomic DNA isolated before immunoprecipitation was also analyzed by PCR and qPCR as the input control. (E) Retro-orbital injection of human recombinant Rln3 to dnStat3-expressing fish (cmlc2:CreER;β-act2:RSdS) for 7-d stimulated cardiomyocyte proliferation at 7 dpa. No detectable effect was observed in wild-type fish (WT) when exposed to the same regimen. A one-time injection of Rln3 to dnStat3-expressing fish at 6 dpa did not significantly increase cardiomyocyte proliferation at 7 dpa. Data are mean ± SEM n = 7–9, *P < 0.05, Student t test, (unpaired, 2-tailed). (F) Confocal images indicating increased cardiomyocyte proliferation in 7 dpa ventricles of animals given seven daily injections of human recombinant Rln3. Brackets indicate injury site. (Insets) Higher-magnification images of the squares; arrows, proliferating cardiomyocytes. (Scale bar, 50 μm.) EXPRESSION / LABELING:
|
|
Expression analysis of Jak1/Stat3 pathway members. (A) In situ hybridization indicating induction of jak1 and stat3 at 1 and 7 dpa with similar dynamics to il6st and socs3b. (Scale bar, 50 μm.) (B) Uninjured and 1-dpa cardiomyocyte RNAs from cmlc2:TRAP fish were processed for qPCR. il11r-α was the most abundantly expressed IL-6 family cytokine receptor in both uninjured and 1-dpa samples. Data are mean ± SEM n = 3. Expression levels were normalized to that of β-actin2, and further normalized to that of uninjured il6r-α. (C and D) qPCR indicating that cntf1 expression levels were decreased and cntf2 expression levels not significantly changed after injury. Data are mean ± SEM n = 3, *P < 0.05, Student t test (unpaired, two-tailed). Expression levels were normalized to that of β-actin2, and further normalized to that of the uninjured sample. (E) qPCR indicating relative expression levels of IL6 family ligands, indicating that il11a is the most abundantly expressed ligand at 1 dpa. Data are mean ± SEM n = 3. Expression levels were normalized to that of β-actin2, and further normalized to that of il6. (F) Sections from 1-dpa fli1:EGFP ventricles were stained with DAPI and imaged for green fluorescence. Then, the same sections were processed for in situ hybridization with an il11a probe. The il11a signal was enriched in and near fli1:EGFP+ nuclei. Arrowheads indicate colocalization between il11a and fli1:EGFP. (Scale bar, 50 μm.) EXPRESSION / LABELING:
|
|
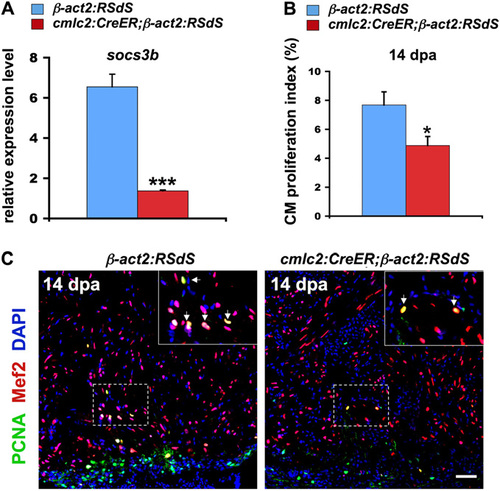
Inhibition of Stat3 signaling in transgenic fish. (A) qPCR indicating reduced expression of the Stat3 target gene socs3b in dnStat3-expressing (cmlc2: CreER; β-act2:RSdS) ventricles at 1 dpa, signifying that endogenous Stat3 activity was impaired. Data are mean ± SEM n = 3, ***P < 0.001, Student t test (unpaired, two-tailed). Expression levels were normalized to that of β-actin2, and further normalized to that of β-act2:RSdS. (B) cmlc2:CreER;β-act2:RSdS and control β-act2:RSdS fish were treated with 4-hydroxytamoxifen (4-HT) at 8 dpa to induce dnStat3 expression, and cardiomyocyte proliferation was analyzed in 14-dpa ventricles. There was an <36% reduction in cardiomyocyte proliferation indices after dnStat3-GFP expression. Data are mean ± SEM n = 5–7, *P < 0.05, Student t test (unpaired, two-tailed). (C) Confocal images indicating reduced cardiomyocyte proliferation in 14-dpa ventricles, after dnStat3-GFP induction at 8 dpa. (Insets) Higher-magnification images of the squares; arrows, proliferating cardiomyocytes. (Scale bar, 50 μm.) EXPRESSION / LABELING:
|
|
Inhibition of Stat3 in cardiomyocytes has no detectable effects during rapid animal growth. (A) Induction of dnStat3-GFP fluorescence in cardiomyocytes after 4-HT incubation in cmlc2:CreER;β-act2:RSdS or control β-act2:RSdS fish, during slow (normal) and accelerated growth conditions. (B) Control (β-act2:RSdS) and cmlc2:CreER;β-act2:RSdS animals each approximately doubled their mass after 10 d of rapid growth conditions. Data are mean ± SEM n = 12. (C) Representative images of cardiomyocyte proliferation using antibodies against PCNA and Mef2, indicating no detectable difference between control (β-act2:RSdS) and dnStat3-expressing fish (cmlc2:CreER; β-act2:RSdS) after 10 d of either slow or rapid growth. Quantified data are in Fig. 3D. (D) Control (β-act2:RSdS) and cmlc2:CreER;β-act2:RSdS embryos were incubated in 4-HT at 4 d postfertilization and allowed to grow to adulthood. Approximately half of the surviving animals from a clutch of 43 fish were cmlc2:CreER; β-act2:RSdS, indicated by myocardial dnStat3-GFP fluorescence. (Scale bars, 50 μm.) |