- Title
-
Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- Authors
- Gupta, V.A., Hnia, K., Smith, L.L., Gundry, S.R., McIntire, J.E., Shimazu, J., Bass, J.R., Talbot, E.A., Amoasii, L., Goldman, N.E., Laporte, J., and Beggs, A.H.
- Source
- Full text @ PLoS Genet.
|
Protein-protein interactions between myotubularin and MTMR12 proteins. (A) GST pull down of MTM1-GST recombinant protein with MTMR12 showed a direct interaction between the two proteins (above). Coomassie blue stained gel (below) showing GST and MTM1-GST protein used in the pull down (B) Co-IP experiments from Cos-1 transfected cells with MTM1 and MTMR12-B10 constructs showing the interaction of the two proteins in the cellular context in-vivo (C) MTMR12 co-immunoprecipitates with myotubularin (using 1G1 monoclonal antibody) in mouse muscle lysates revealed with anti-MTMR12 polyclonal antibody (upper panel) and with the 2827 polyclonal anti-myotubularin (bottom panel). (D) Confocal microscopic immunofluorescence studies of longitudinal frozen sections of skeletal muscle from mouse tibialis anterior muscle with sarcomeric markers. Individual immunostaining with 2827 anti-myotubularin or anti-MTMR12 showed similar striated localization in skeletal muscle (top panel). Double-immunostaining with both proteins showed a co-localization of myotubularin and MTMR12 to Triads and partial co-localization with the ryanodine receptor, RyR1, a sarcoplasmic reticulum marker, but not with α-actinin, a Z-line marker. Scale bar = 10 μm. |
|
Expression patterns and morpholino-based knockdown of mtmr12 in developing zebrafish. (A) Whole mount in-situ hybridization detected ubiquitous expression of mtmr12 and mtm1 transcripts in zebrafish embryos at 1 dpf (above). Below is RT-PCR analysis of mtm1 and mtmr12 expression during zebrafish development using RNA extracts from whole zebrafish embryos at indicated developmental timepoints. (B) Synergistic expression level of Mtm1 and Mtmr12 transcripts and protein at indicated time points of C2C12 differentiation (0–9 days) monitored by RT-quantitative PCR (corresponding histogram, *Pd0.05) and by western blot analysis (right panel). (C) Live embryos at 3 dpf injected with control, mtmr12, mtm1 or both mtmr12 and mtm1 morpholinos in normal (left) and polarized lights (right). mtmr12 morphant fish showed a dorsal curvature in skeletal muscle and reduced birefringence in polarized light similar to mtm1 morphant embryos. mtmr12 morphant fish also exhibited pericardial edema (arrow). mtmr12-mtm1 double knockdown fish exhibited smaller size and reduced birefringence relative to mtm1 or mtmr12 alone morphant fish. (D) mtmr12 mRNA levels in mtmr12 morphant zebrafish following injection of two different amounts of morpholino (indicated below, upper panel). In mtm1 morphant fish, no residual myotubularin was observed showing that mtm1 morpholinos are completely penetrant to the limits of detection for western blotting. (E) Over-expression of human MTMR12 mRNA rescued small body length and skeletal muscle abnormalities observed in mtmr12 morphant embryos. (F) Quantification of the chorion hatching at 60 hpf. The number of embryos was quantified in three independent clutches (number of embryos in each clutch = 90–120). (G) Quantification of touch evoke response at 3 dpf (n = 5–8 embryos were assayed in each morpholino group).*Pd0.01. PHENOTYPE:
|
|
Abnormal histology of MTMR12-deficient zebrafish. (A)Toluidine blue stained longitudinal sections of skeletal muscle in control and morphant fish at 3 dpf. In comparison to the control fish, mtmr12 morphants showed disorganized myofibers (arrowhead) with central nucleation (arrow), similar to histological changes observed in the skeletal muscle of mtm1 morphant fish (arrow). Knockdown of both mtm1 as well as mtmr12 results in severe muscle disorganization greater than seen in mtm1 or mtmr12 alone morphants. (B) Centrally nucleated myofibers were quantified. Serial sections from 3–4 different embryos were analyzed and the relative number of centrally nucleated fibers in the middle somites (10–13) were counted. (C) Hematoxylin and Eosin staining of mtmr12 morphant zebrafish at different time points. An increase in sarcomeric disorganization was observed at 3 dpf in comparison to 2 dpf in mtmr12 morphants (arrow) (D) Centrally nucleated myofibers were quantified. Serial sections from 6 different embryos were analyzed and the relative number of centrally nucleated fibers in the middle somites (10–13) were counted. Scale bar = 10 μm. PHENOTYPE:
|
|
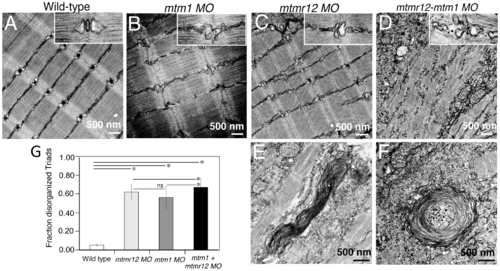
Ultrastructural abnormalities in MTMR12 deficiency. Transmission electron micrographs of skeletal muscles in control and morphant fish. In comparison to wild-type controls (A), mtmr12 morphant fish displayed abnormal triads (C, high magnification, inset). mtm1 morphant muscle also had absent or disorganized triads (B). mtmr12-mtm1 double knockdown morphants displayed exacerbated defects in muscle with many absent Z-lines in addition to disorganized triads (D inset shows high magnification view). Higher magnification examination showed whorled membranous structures in mtmr12 and mtmr12-mtm1 morphants (E–F). (G) Histograms represent quantification of disorganized triads of the single or double morphants. Total number of triads were counted in at least 15 myofibers within each embryo (n = 5 embryos). *Pd0.05, ns: statistically not significant. PHENOTYPE:
|
|
Myotubularin and PtdIns3P alterations in mtmr12 morphants. (A) Immunofluorescence of control and mtmr12 knockdown fish showed significantly decreased myotubularin staining in mtmr12 knockdown fish in images taken under identical conditions. Immunofluorescence detection of PtdIns3P showed apparent increases of this myotubularin substrate in mtmr12 morphant embryos as compared to controls. (B) PtdIns3P levels are increased in mtmr12, mtm1 and mtm1-mtmr12 morphant zebrafish, *Pd0.05. Total lipids were extracted from zebrafish at 3 dpf and PtdIns3P levels were measured using a lipid-protein overlay enzyme-linked immunosorbent assay. EXPRESSION / LABELING:
|
|
Loss of protein stability in the absence of myotubularin-MTMR12 interactions. (A) Knockdown of mtmr12 resulted in a strong decrease of myotubularin protein in mtmr12 morphant zebrafish at 3 dpf by western blotting. Western blot analysis was done on three independently injected clutches (n in each clutch = 50–75). The histogram at right shows normalized amounts of myotubularin in control and mtmr12 morphant zerbrafish, *Pd0.01. (B) siRNA-mediated knockdown of Mtmr12 in C2C12 myoblasts leads to decreased protein levels of MTM1. Histograms showed western blot quantification of MTM1 and MTMR12 with reference to α-actinin as a loading control. Data represent mean of 3 independent experiments, * P<0.05. (C) Mtmr12 siRNA treated myoblasts were differentiated into myotubes and tested for protein expression of MTM1, MTMR12, desmin and myogenin. Mtmr12 knockdown in myotubes leads to decreased protein levels of MTM1 and increased amounts of the intermediate filament protein desmin, but do not affect myogenin levels. α-Actinin was used as the loading control (histograms). Data represent mean of 3 independent experiments, * P<0.05. (D) Labeling of α-actinin and desmin in C2C12 myoblast and myotubes treated with Mtmr12 siRNA or scramble control siRNA. Mtmr12 knockdown cells showed abnormal accumulation of desmin in both stages (arrow). (E) Quantification of myotubes at 2, 4, 6 and 9 days of differentiation showed no significant differences between siRNA-Mtmr12 cells and scramble siRNA. Data were obtained from 2 independent experiments (* P<0.05) and minimum of 100 cells per condition were counted. (F) Mtm1 knockout mice exhibited highly reduced levels of MTMR12 protein in skeletal muscle at pre-symptomatic (2 weeks) as well as symptomatic stages (5 weeks). The histogram on right shows normalized amounts of MTMR12 in control and Mtm1KO Mice, *Pd0.05. (G) siRNA-based Mtm1 knockdown in C2C12 cells led to no reduction in MTMR12 protein as seen by western blot analysis. β-actin/GAPDH were used as loading controls in western blots. The histogram at right shows normalized amounts of MTMR12 in control and Mtm1 knockdown cell line, *Pd0.05. EXPRESSION / LABELING:
|
|
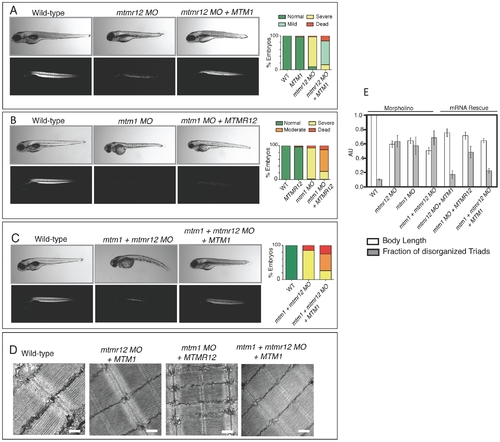
Rescue of mtmr12 morphant phenotypes by MTM1. The ability of human MTM1 or MTMR12 transcripts to rescue abnormalities seen in morphant zebrafish was classified in to phenotypic index of five groups: Normal, mild, moderate, severe and dead, described in the table depending on body length, birefringence and ultrastructure of skeletal muscle. (A) Polarized light microscopy of 3 dpf live embryos showed that birefringence of mtmr12 morphant embryos increased significantly upon overexpression of human MTM1 mRNA. (B) Overexpression of human MTMR12 mRNA in mtm1 morphant fish resulted in a mild rescue of skeletal muscle defects as seen by birefringence of zebrafish embryos. (C) Overexpression of human MTM1 mRNA in mtm1-mtmr12 morphant fish resulted in a moderate rescue of skeletal muscle defects as seen by birefringence of zebrafish embryos. (D) Electron microscopy showed normal skeletal muscle structure of mtmr12 and mtm1-mtmr12 morphant fish rescued with MTM1 mRNA but displayed disorganized triads in mtm1 morphants that were rescued with MTMR12 mRNA. (E) Quantification of the body length and disorganized triads in morphant and rescued fish. Body length was measured in 10–15 embryos in each group. Total number of triads were counted in at least 15 myofibers within each embryo (n = 5 embryos). Pd0.05. PHENOTYPE:
|
|
Myotubularin-MTMR12 interactions in XLMTM. (A) Schematic diagram of different domains of myotubularin protein displaying representative pathogenic mutations found in XLMTM patients or an artificial inactivating mutation C375S* (GRAM, N terminal lipid or protein interacting domain; RID, putative membrane targeting motif; PTP/DSP, phosphatase domain; SID, protein-protein interacting domain; CC, coiled-coil domain; PDZB, PDZ binding site). (B) Wild-type or mutant MTM1-B10 fusion proteins with indicated missense mutations and wild-type MTMR12-GFP proteins were overexpressed in Cos1 cells. Immunoprecipitation of protein extracts with anti-B10 tag antibody showed that mutations on GRAM or RID domains disrupt the interactions between MTM1 and MTMR12. (C) Western blotting of XLMTM patient myotubes showed that mutants that decrease the stability of myotubularin protein also results in a reduction of MTMR12 levels. Histograms depict the western quantification for panels (B) and (C). Asterisks indicate statistically significant differences from measurements of wild type controls, Pd0.05. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|