- Title
-
Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish
- Authors
- Huitema, L.F., Apschner, A., Logister, I., Spoorendonk, K.M., Bussmann, J., Hammond, C.L., and Schulte-Merker, S.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
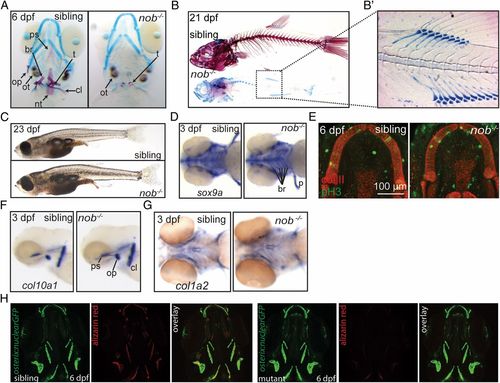
Nob mutants lack a mineralized skeleton. (A) Alizarin red/alcian blue staining of sibling and mutant nob embryos. Cartilage elements appear normal in mutants. All bone is absent, but teeth and otoliths are present. (B) Skeletal staining of 21-dpf sibling and nob mutant individuals. (B2) Enhanced contrast image highlighting the correctly patterned but unmineralized vertebra anlagen. (C) Images of sibling and mutant nob fish at 23 dpf, demonstrating that nob mutants are indistinguishable from siblings at the gross morphological level. (D) Whole-mount in situ hybridization detecting the chondrogenic marker sox9a. (E) Confocal projection of Meckel’s cartilage of a sibling versus mutant embryo showing no difference for anti-type II collagen (red) or the proliferation marker anti–phospho-Histone H3 (green). (F and G) Whole-mount in situ hybridization detecting the osteoblast markers col10a1 (F) and col1a2 (G) in 3-dpf nob mutant and sibling embryos. (H) Osterix:GFP expression, marking early osteoblasts in 6-dpf nob mutant and sibling embryos. br, fifth branchial arch; cl, cleithrum; nt, notochord tip; op, operculum; ot, otolith; ps, parasphenoid; t, teeth. EXPRESSION / LABELING:
PHENOTYPE:
|
|
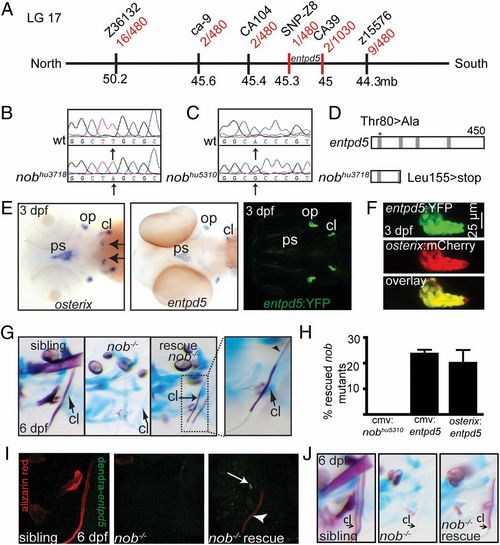
Nob mutants encode entpd5, which is specifically expressed in osteoblasts. (A) Meiotic map of the nobhu3718 locus. Recombinants per total number of mutants tested for each polymorphic marker are depicted in red, and markers used for mapping are in black. (B) Sequencing of nobhu3718 revealed a Leu155>stop mutation. (C) Sequencing of nobhu5310 revealed a Thr80>Ala mutation transversion. (D) Schematic representation of the predicted protein structures for Entpd5 and nobhu3718. The asterisk indicates the position of the mutation in nobhu5310. Gray bars, apyrase domains within Entpd5 (35). See also Fig. S1. (E) (Left and Center) Entpd5 and osterix are almost identically expressed in 3-dpf embryos, with osterix expression in the region of future teeth (arrows) being the single exception at this stage. (Right) Representative image of a 3-dpf entpd5:YFP transgenic embryo showing an expression pattern that matches endogenous entpd5 transcript distribution (Center). (F) Coexpression of entpd5:YFP and osterix:mCherry in the operculum of 3-dpf embryos. (G and H) entpd5 cDNA injection (CMV promoter) into nobhu3718 mutants results in mosaic mineralization (G). Arrows point at cleithra in G; the arrowhead points to a nonmineralized osteoid in a partially mineralized cleithrum. Bar graphs in H represent percentages of rescued mutant embryos at 6 dpf (expressed as mean ± SEM, three independent experiments). (I) Alizarin red staining of a mutant nob embryo that had been injected with a cmv:dendra-t2a-entpd5 expression construct at the one-cell stage. The cleithrum is partially mineralized in the mutant (arrowhead); however, there are no dendra-positive cells immediately abutting the mineralized structure (arrow). (J) Representative example of an embryo partially rescued by endothelial-specific expression of entpd5 (kdr-like:entpd5). EXPRESSION / LABELING:
|
|
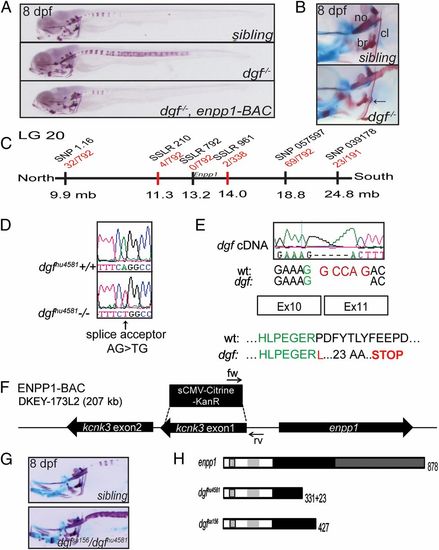
Ectopic mineralization phenotypes in dgf mutants. (A) Skeletal staining of a sibling, a dgf mutant, and a dgf mutant rescued by an enpp1 BAC transgene. Dgf mutants exhibit axial hypermineralization, resulting in partially fused vertebral centra. (B) Hypermineralization also affects the cleithra, which in dgf mutants typically exhibit nodule-like protrusions (arrow). no, notochord. (C) Meiotic map of the dgf locus. Recombinants per total number of mutants tested for each polymorphic marker are depicted in red, and markers used for mapping are in black. (D) Sequencing of dgfhu4581 revealed a mutation in the splice acceptor site before exon 11 of the enpp1 gene. (E) The consequence at the transcript level is a deletion of 5 bp (Upper), resulting in a translational frameshift and a predicted stop codon after 23 amino acids (Lower). (F) Schematic representation of the BAC construct generated for the transgenic rescue of the dgf4581 phenotype. Two genes are contained on the BAC, and kcnk3 was inactivated through a recombineering approach. (G) Dgfsa156 fails to complement dgfhu4581. (H) Schematic drawing of the predicted protein forms of enpp1, dgfhu4581, and dgfsa156, respectively. Enpp1 is a type II transmembrane protein. Striped box, transmembrane domain; light gray, somatomedin B-like binding domains; black, catalytic domain; dark gray, nuclease-like domain (36). See also Fig. S2. PHENOTYPE:
|
|
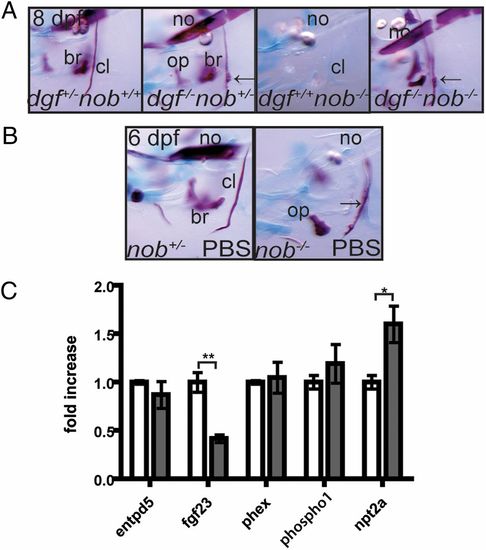
Phosphate homeostasis is disturbed in nob mutants. (A) Nob mutants can mineralize their skeleton in the absence of dgfhu4581 function, and will even show ectopic mineralization (arrows point at protrusions of the cleithrum, a typical feature of dgf mutants). (B) Exogenous phosphate partially rescues the nob mutant phenotype. Arrow points at partially mineralized cleithrum. All images represent lateral views of the head skeleton at 6 dpf. (C) Quantitative PCR on 7-dpf embryos revealed that, compared with siblings (white bars), fgf23 expression levels are significantly down-regulated and npt2a expression is significantly up-regulated in nob mutant embryos (gray bars). Results are expressed as mean ± SEM of three independent experiments, with *P < 0.05 and **P < 0.005. |
|
(A) Alcian blue staining is unaltered in nob mutants versus siblings. The images are similar to the ones presented in Fig. 1A, but taken with higher contrast settings to allow an appreciation of normal chondrocyte morphology in mutant embryos. (B) Multiple sequence alignment of Entpd5 proteins demonstrating the conserved nature of zebrafish Thr80 in the first apyrase domain. |
|
dgf-/- mutant embryos can be rescued by transgenic insertion of a BAC containing the enpp1 gene, and dgfsa156 encodes a mutation in enpp1. (A) BAC-rescued embryos exhibit a wild-type phenotype (shown in Fig. 3A). The SNP variant (arrow) is linked to the dgf mutation (sequence, Middle). (B) PCR indicating the presence of the enpp1 BAC in rescued dgf-/- embryos and its absence in nonrescued dgf-/- embryos. (C) Sequencing of the dgfsa156 allele, kindly provided by The Sanger Center, reveals an Arg427>stop in the enpp1 gene. |

Unillustrated author statements PHENOTYPE:
|