- Title
-
Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant persephone
- Authors
- Hailey, D.W., Roberts, B., Owens, K.N., Stewart, A.K., Linbo, T., Pujol, R., Alper, S.L., Rubel, E.W., and Raible, D.W.
- Source
- Full text @ PLoS Genet.
|
Phenotype of the persephone mutant. (A) Hair cell protection in homozygous persephone mutants. 5 dpf zebrafish (progeny of a heterozygous incross) were treated with or without 200 μM neomycin for 30 min, and then rinsed. After 1 hr recovery in fresh embryo media, hair cells were labeled with the vital dye DASPEI. Left panels are differential interference contrast (DIC) images and right panels are corresponding fluorescent images. Top, wildtype sibling (+/+), mock-treated, Middle, wildtype sibling (+/+), treated with 200 μM neomycin, and Bottom, persephone homozygote treated with 200 μM neomycin. Red arrows indicate examples of neuromasts present in the untreated wildtype and treated persephone larvae. Mutant larvae show dramatic retention of hair cells relative to their siblings. (B) Parvalbumin antibody staining of hair cells in representative fish from an in-cross of persephone heterozygotes. Wildtype siblings show loss of hair cells when treated with 200 μM neomycin. Homozygous persephone mutants show a dramatic retention of parvalbumin-stained hair cells following neomycin treatment, consistent with DASPEI hair cell staining results. (C) Protection observed in persephone is not due to a delay in neomycin-induced hair cell death. persephone mutants (blue line) exposed to neomycin for 1 hr, rinsed and maintained for 6 or 24 hr in fresh EM, and then assessed for hair cell death, do not show significantly greater hair cell death than those assayed after 1 hr. Hair cell death is not delayed in persephone. Wildtype siblings treated in parallel (green line) are shown for comparison. (n = 10 fish, 10 neuromasts per fish; Error bars: S.D.; p value<0.001). |
|
persephone hair cells have normal morphology and show only modest ultrastructural changes when treated with neomycin. Zebrafish (5 dpf) treated with or without neomycin were euthanized, tails were removed for genotyping, and heads were fixed for transmission electron microscopy. Images of transverse sections through neuromasts are shown. (N: hair cell nuclei, m:mitochondria, sc: support cell, k: kinocilia. Top panels A–C and bottom panels G–I show hair cells from wildtype siblings. Top panels D–F and bottom panels J–L show hair cells from persephone homozygotes. Panels were either mock-treated or exposed to 50 µM or 200 µM neomycin. Hair cells of mock-treated wildtype siblings and persephone mutants show organized stereocilia, a large central nucleus, and normal mitochondrial morphology. There are no readily apparent differences between the wildtype siblings and persephone mutants. Hair cells of persephone mutants treated with 50 μM neomycin show either mild mitochondrial swelling (E,K) or are indistinguishable from mock-treated wildtype siblings. In comparison wildtype siblings (B,H) show swollen mitochondria and fused stereocilia. Most neuromasts from wildtype siblings treated with 200 µM neomycin lack hair cells, and remaining hair cells exhibit severe damage including condensed nuclei and severely swollen mitochondria or signs of cytolysis (C,I). Hair cells of persephone mutants treated with 200 μM neomycin show minimal damage. Stereocilia are typically oraganized and intact; some but not all hair cells show modestly swollen mitochondria (F,L). See Table 1 for quantification of neomycin-induced damage across genotypes. Top scale bar = 5 μm; Bottom scale bar = 1 μm. PHENOTYPE:
|
|
Mutant Slc4a1b is mislocalized and fails to transport chloride. (A) Confocal images of zebrafish embryos transiently expressing wildtype Slc4a1b–GFP. mRNA was injected into one cell stage embryos. Cells along the anterior dorsal aspect of embryos were imaged at 48 hpf. Wildtype protein is localized to the plasma membrane. B) Confocal images of zebrafish embryos expressing mutant Slc4a1b(S298F)-GFP. Slc4(S298F)-GFP is present in intracellular vesicles, with complete loss of plasma membrane localization. (C) Chloride influx into Xenopus laevis oocytes expressing either zebrafish Slc4a1b (zSlc4a1b) or the closest human homolog, SLC4A1 (hSlc4a1). Zebrafish Slc4a1b transports radiolabeled chloride across oocyte membranes as efficiently as its nearest human homolog. This activity is blocked by DIDS (200 μM), an inhibitor of Slc4 family-mediated anion exchangers. (D) Comparison of chloride influx into Xenopus laevis oocytes expressing wildtype or mutant (S298F) zSlc4a1b, or wildtype zSlc4a1b-GFP. zSlc4a1(S298F), the persephone gene product, exhibits complete loss of chloride influx activity. Notably, GFP-tagged wildtype slc4a1b exhibits no significant reduction of chloride transport activity. (n indicated above each sample; Error bars = SEM). |
|
persephone, SLC4 inhibitors, and high bicarbonate all reduce gentamicin uptake. (A) Gentamicin-Texas Red (GTTR) labeling is shown in neuromasts in a wildtype sibling (top panels) and persephone mutant (bottom panels). Top down and cross sectional views are shown with identical image adjustments made to all GTTR channel images. persephone mutants show dramatically reduced intracellular GTTR signal. (B) Quantified GTTR signal after a 3 min exposure is reduced in persephone mutants compared to homozygous wildtype and heterozygous siblings. Free-swimming fish were exposed to GTTR for 3 min and rinsed by basket transfer. Neuromasts of individual fish were immediately imaged and total fluorescence signal from GTTR was quantified as described in Materials and Methods and Figure S3. After imaging, fish were individually genotyped by dCAPS. (n≥10 larvae per group, 3 neuromasts per larvae. Error bars: S.D., p value: 0.003 for difference in means between wildtype siblings and persephone larvae.) (C) SITS, DIDS and high bicarbonate reduce hair cell GTTR uptake. For all conditions: free-swimming wildtype 5dpf fish were treated 1 hr with either vehicle carrier, 200 μM SITS or DIDS, or 10× [bicarbonate] buffered with HEPES, and then exposed for 3 min to GTTR as in (B). GTTR uptake is significantly reduced in fish treated with concentrations of SITS, DIDS, and bicarbonate that protect hair cells from neomycin exposure. (n = 10 fish, 3 neuromasts per fish. Error bars: S.D. p value: <0.0001 for difference in means between 0 and 200 μM SITS, 0 and 200 μM DIDS, and 1× and 10× bicarbonate). PHENOTYPE:
|
|
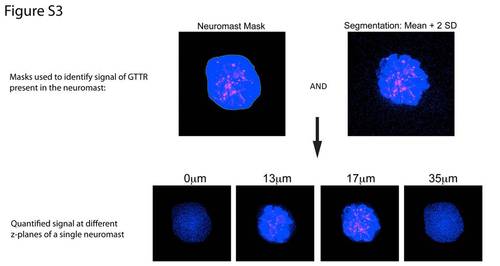
Quantification of GTTR signal. To quantify GTTR signal, 3-D image stacks were collected of entire neuromasts at z-increments of 0.4 μm. Total fluorescence of neuromasts was calculated in individual neuromasts using 3i Slidebook Image Analysis software. A mask was drawn around visually identified neuromasts. The image on the top left shows a mask outlining a neuromast. Pixels within this mask are shown in blue; GTTR signal is red. A second identical mask was placed outside the neuromast to quantify background. Neuromast and background signals were segmented to discard points less than 2 standard deviations above the mean background value. The pixels that meet this criteria are shown in blue in the top right image. These two masks were then combined to generate a mask (the signal mask) that contained the region of the neuromast and required that values in that the mask be at least the mean value of the background plus two standard deviations. The bottom panels show this mask at different z-planes for the neuromast shown in the top panel. Blue points represent those points included in the mask. We calculated the mean intensity of the blue points in the signal mask. This mean value of the neuromast signal was then divided by the mean background signal to yield the relative intensity of the neuromast signal relative to background. |