- Title
-
Effective gene trapping mediated by sleeping beauty transposon
- Authors
- Song, G., Li, Q., Long, Y., Gu, Q., Hackett, P.B., and Cui, Z.
- Source
- Full text @ PLoS One
|
Inducible activity of the tilapia Hsp70 promoter (TiHsp70) at 37 °C. (A) A novel gene-trap vector mediated by Sleeping Beauty transposon. IR/DR(L) and IR/DR(R), left and right inverted repeat/directed repeat of the SB transposon; SA, splice acceptor; IRES, internal ribosome entry site; Neo, kanamycin resistance gene; poly(A), poly(A) signal; TiHsp70, tilapia Hsp70 promoter; SB11, SB11 transposase gene. (B) EGFP was used to monitor the inducible activity of TiHsp70 in vitro and in vivo. HeLa cells were transfected with pTiHsp70-EGFP at the density of about 80% confluence, treated in medium at 37°C for 1 h and recovered at 32°C for 2 h after transfection. Images were taken under a Nikon TE2000 fluorescent microscope and cell numbers in three fields of view were counted. Zebrafish embryos at one-cell stage were microinjected with pTiHsp70-EGFP. Injected embryos at 24 hpf were incubated in rearing water at 37°C for 1 h and then recovered at 28°C for 2 h. Low magnification fluorescent imaging of zebrafish embryos was performed on a SteReo Lumar V12 microscope form Zeiss and total embryos in three dishes were counted. (C) Statistical analysis of EGFP-expressing cells or embryos in (B). Data are given as means ± standard Deviation (n = 3). * indicate P<0.05 versus the corresponding control. (D) Western blot analysis of EGFP in HeLa cells and zebrafish embryos. Heatshock-treated and -untreated cells (transfected with pTiHsp70-EGFP) and embryos (injected with pTiHsp70-EGFP) samples were undergone western blot and the pEGFP-F plasmid was used as a positive control. The expression of reference gene β-actin was also analyzed in these samples. |
|
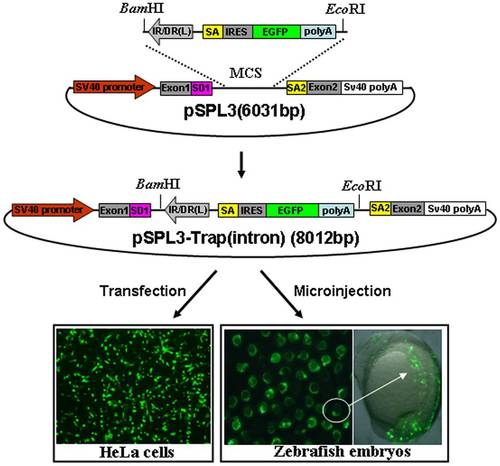
Activity of the trapping cassette in an intron of pSPL3 vector. The trapping cassette was subcloned into the BamHI/EcoRI site of pSPL3 vector to generate the pSPL3-Trap(intron), which was used for transient transfection of HeLa cells at 80% confluence. Images were taken under a Nikon TE2000 fluorescent microscope at 48 h after transfection and cell numbers from three independent transfections were counted. Zebrafish embryos at one-cell stage were microinjected with the pSPL3-Trap(intron). Injected embryos at 24 hpf were imaged under a SteReo Lumar V12 microscope form Zeiss and total embryos in three dishes were counted. The ectopic expression of EGFP in one embryo was enlarged and shown in a merged image. SD1, splice donor for exon1; IR/DR(L) and IR/DR(R), left and right inverted repeat/directed repeat of the SB transposon; SA, splice acceptor; IRES, internal ribosome entry site; EGFP, enhanced green fluorescence protein gene; poly(A), poly(A) signal; SA2, splice acceptor for exon2. |
|
Activity of the trapping cassette in an exon of pSPL3-E3 vector. pSPL3-E3 was generated by insertion of an exon from carp beta-actin gene (Exon3). The trapping cassette was then sucloned into the BamHI/EcoRI site of pSPL3-E3 vector to generate the pSPL3-E3/Trap(exon), which was used for transient transfection of HeLa cells at 80% confluence. Images were taken under a Nikon TE2000 fluorescent microscope at 48 h after transfection and cell numbers in three independent transfections were counted. Zebrafish embryos at one-cell stage were microinjected with the pSPL3-E3/Trap(exon). Injected embryos at 24 hpf were imaged under a SteReo Lumar V12 microscope form Zeiss and total embryos in three dishes were counted. The ectopic expression of EGFP in one embryo was enlarged and shown in a merged image. SD1, splice donor for exon1; SA3, splice acceptor for exon3; IR/DR(L) and IR/DR(R), left and right inverted repeat/directed repeat of the SB transposon; SA, splice acceptor; IRES, internal ribosome entry site; EGFP, enhanced green fluorescence protein gene; poly(A), poly(A) signal; SD3, splice donor for exon3; SA2, splice acceptor for exon2. |
|
An IRES sequence is required for independent expression of the reporter gene. (A) The ECMV/IRES element in the pSPL3-Trap(intron) vector was deleted to generate the pSPL3-Trap(intron)ΔIRES. SD1, splice donor for exon1; IR/DR(L) and IR/DR(R), left and right inverted repeat/directed repeat of the SB transposon; SA, splice acceptor; IRES, internal ribosome entry site; EGFP, enhanced green fluorescence protein gene; poly(A), poly(A) signal; SA2, splice acceptor for exon2. (B) pSPL3-Trap(intron) and pSPL3-Trap(intron)ΔIRES constructs were transfected into HeLa cells at 80% confluence, respectively. Images were taken under a Nikon TE2000 fluorescent microscope at 48 h after transfection and cell numbers in three independent transfections were counted. (C) Zebrafish embryos at one-cell stage were microinjected with the pSPL3-E3/Trap(exon). Injected embryos at 24 hpf were imaged under a SteReo Lumar V12 microscope form Zeiss and total embryos in three dishes were counted. (D) Statistical analysis of EGFP-expressing cells in (B) or embryos in (C). Each construct was tested three times and each experiment was done in triplicate. Data are given as means ± standard Deviation. ** indicate P<0.01 versus the corresponding control. |
|
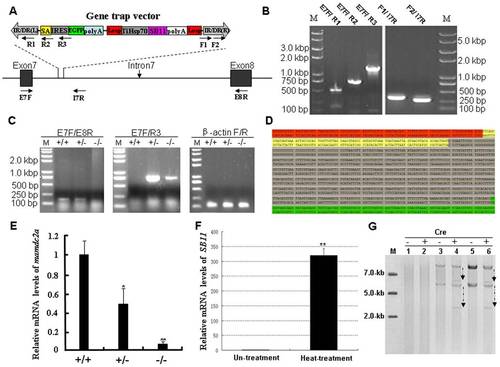
Molecular analysis of the trapped gene mamdc2a in transgenic zebrafish. (A) Schematic representation of the insertion of gene trap cassette in the intron7 of gene. The locations of primers used for molecular analysis are indicated. (B) Integrity analysis of gene trap cassette in transgenic fish. Genomic DNA was extracted from the trail of transgenic fish and subjected to PCR analysis with primes indicated in (A). (C) RT-PCR analysis of mamdc2a transcript in wild-type (+/+), heterozygous (+/-) and homozygous (-/-). Primers E7F/E8R were used to amplify the endogenous transcript and primers E7F/R3 were used to amplify the exon7-EGFP fusion transcript. The β-actin transcript was amplified as an internal control. (D) The fusion transcript generated from the proper splicing of EGFP gene and its upstream exon in mamdc2a gene. Sequences in red represent exon7 of mamdc2a gene. Sequences in yellow represent the SA signal. Sequences in gray indicate the IRES followed by the partial EGFP sequence in green. (E) qRT-PCR was performed with primers E7F/E8R to determine the expression of endogenous mamdc2a gene in wild-type, heterozygous and homozygous embryos at 72 hpf. The mamdc2a levels were normalized to the β-actin mRNA levels. (F) qRT-PCR analysis of SB11 expression in 24 hpf embryos from mamdc2a-transgenic line after heat shock at 37°C for 3 h and recovery at 28°C for 2 h. (G) Detection of Cre-mediated TiHsp-SB11 excision by southern hybridization. Cre reombinase mRNA was microinjected into wild-type, heterozygous and homozygous embryos. “+” represents the presence of Cre recombinase and “-” represents the absence of Cre recombinase. Line1–2: DNA from wild-type. Line 3–4: DNA from heterozygous. Line 5–6: DNA from homozygous. |