- Title
-
Cdc42 GTPase and Rac1 GTPase act downstream of p120 catenin and require GTP exchange during gastrulation of zebrafish mesoderm
- Authors
- Hsu, C.L., Muerdter, C.P., Knickerbocker, A.D., Walsh, R.M., Zepeda-Rivera, M.A., Depner, K.H., Sangesland, M., Cisneros, T.B., Kim, J.Y., Sanchez-Vazquez, P., Cherezova, L., Regan, R.D., Bahrami, N.M., Gray, E.A., Chan, A.Y., Chen, T., Rao, M.Y., and Hille, M.B.
- Source
- Full text @ Dev. Dyn.
|
p120 catenin δ1 is the predominant p120 catenin mRNA in early developing embryos. Real-time PCR was used to make a quantitative analysis of relative expression levels of p120 catenin δ1, p120 catenin δ2a, and ARVCF at various stages from 500-cell to 12-somite embryos of developing zebrafish. Relative amounts were calculated using the 2-ΔΔC(t) method to standardize target gene expression to an endogenous standard (EF-1α). Plotted values represent the mean of all 2Standarized ΔC(t) for a developmental stage and primer combination, ± 2 s.e.m. See Supp. Figure S1 for more details. For most stages and primer combinations N = 6. EXPRESSION / LABELING:
|
|
Two isoforms of p120 catenin proteins are detected in homogenates of embryos between 3.7 and 38 hpf in SDS-PAGE Western blots using a polyclonal antibody to p120 catenin. SDS-PAGE homogenates from 3 embryos were added to each lane. Stages shown are: oblong about 4,000 cells (3.7 hpf); shield (6 hpf); bud at the end of epiboly (11 hpf); 2-somite stage (11 hpf); 10-somite stage (14 hpf); 21-somite stage (19.5 hpf); Prim 5 (24 hpf); Prim 25 (38 hpf). The p120 catenin protein that is initiated at Met-1 has a Mr of 120 k, and the protein initiated at Met-3 has a Mr of 100 k. B: Injection of antisense splice morpholino oligonucleotides (Sp-MO) blocks new mRNA synthesis and decreases synthesis of 100-kDa p120 catenin proteins initiating at Met-3. Zebrafish embryos were collected at the 1-cell stage and injected with 6 or 8 ng of δ 1 splice-MO. Zebrafish embryos were counted, and quantitatively lysed in SDS-PAGE buffer as described in the Experimental Procedures section resulting in the loading of 1-embryo per lane. Uninjected embryos were homogenized at shield (6 hpf), 80 to 90% epiboly (9 hpf), and bud (10 hpf) and δ1 splice-MO-injected zebrafish embryos were homogenized at shield and bud. All samples were analyzed by SDS-PAGE Western blot with a polyclonal anti-p120 catenin antibody. C: Abundance of p120 was measured as described in the Experimental Procedures section and plotted on a bar graph. The Y-axis represents relative density in arbitrary units determined by ImageJ with a background correction using Rolling Ball Subtraction. |
|
Knockdown of p120 catenin δ1 with the splice-site morpholino led to narrow, extended somites and knobby notochord protrusions, which were rescued by co-injection of Xenopus p120 catenin mRNA (Xp120 catenin mRNA). A: Micrographs of dissected sibling embryos in dorsal view, anterior to the top right, analyzed with in situ hybridization using a myoD probe at the 9- to 10-somite stage. Embryos were injected as indicated in the panels with buffer, δ1 splice-MO (Sp-MO) and/or Xenopus p120 catenin δ1 mRNA (Xp120 mRNA). Scale bars = 100 μm. B: Summary of the average width of the second-to-last somite of in situ hybridized embryos using a myoD probe at the 5- to 13-somite stages. Embryos were injected as in A. C: Summary of the average width-to-length ratio of the second-to-last somite of embryos from 4-somites to 13-somites. Error bars in B and C represent s.e.m. The number of 2 to 35 embryos is marked on the bars. PHENOTYPE:
|
|
Dorsal view, anterior to the top right, of dissected sibling embryos at the 4- and 7-somite stages analyzed with in situ hybridization using myoD to mark somites and presumptive slow muscle cells and a papc probe to mark presomitic mesoderm. The lateral edges of the papc staining of the presomitic mesoderm are marked with arrows in at top left. One can compare Figures 3A and 4A to distinguish the myoD and papc domains. Embryos were uninjected or injected with δ1 splice-MO (Sp-MO). Scale bars = 100 μm. B: Summary of the average width of the tailbud at the end of the notochord as marked with black arrows in A at the 1- to 9-somite stages for similar injections. Error bars represent s.e.m; numbers of embryos indicated on each bar. |
|
p120 catenin δ1 depletion by δ1 splice-MO causes severe defects in somite formation in later stages of development including defects in yolk extension, tail extension, and eye formation. Embryos were imaged with Nomarski optics. A: Images of side views of live zebrafish embryos, anterior left, at the 14-somite stage (16 hpf). Embryos were uninjected or injected as indicated in the panels with δ1 splice-MO (Sp-MO) and/or Xenopus p120 catenin δ1 mRNA (Xp120 mRNA). Ventral arrows indicate the positions of the head and tailbud. Scale bars = 250 μm. B: Embryos were injected with 6 pg δ1 splice-MO (Sp-MO). C: Comparisons of side views of embryos that were uninjected or injected with δ1 splice-MO and/or Xp120 catenin δ1 mRNA at the 1-cell stage, and then imaged at the Prim 15 stage (30 hpf, anterior top). Eighty-six percent of the embryos injected with Sp-MO survived to Prim 15. Scale bars = 160 μm. PHENOTYPE:
|
|
WT-Cdc42 is downstream of p120 catenin δ1 during axis extension in zebrafish embryos. A: Representative 10-somite embryo side views, dorsal up, anterior left of embryos uninjected, injected with δ1 splice-MO (Sp-MO) and/or WT-Cdc42. Scale bars = 100 μm. B–D: A significantly greater portion of p120 catenin δ1 splice-MO-treated embryos recover their normal phenotype when co-injected with WT-Cdc42. Shown are the percentage of embryos restored to normal (uninjected) phenotypes by co-injection of Sp-MO with (B) WT-Cdc42 mRNA, (C) CA-Cdc42 mRNA, or (D) DN-Cdc42 mRNA. Also shown are the effects of injecting the mRNAs alone. E, F: The decrease in axis extension in 3- to 10-somite embryos as indicated by an increase in the head-to-tail distance. Distances are normalized to the uninjected embryos (clear bar), which were set at 100% for each different clutch of eggs. Embryos were injected at the 1-cell stage with 6 ng Sp-MO (light grey box). These baseline measurements are followed by measurements on embryos co-injected with 6 ng Sp-MO and 1 pg mRNA (grey boxes) or 1 pg mRNA alone (black boxes). The types of Cdc42 mRNAs injected and the stage of the embryos are indicated above the bars. The numbers of embryos observed are given on the graph bars. |
|
Wild-type Rac1 mRNA (WT-Rac1 mRNA) rescues 90% of the &dger1 splice-MO knockdown of p120 catenin δ1. A: Representative 16-somite embryos side views, dorsal up, anterior left, uninjected, and injected with δ1 splice-MO (Sp-MO) and WT-Rac1 mRNA as indicated. B: Summary of the distance between the head and tailbud as marked with arrows in A. C: Side views with dorsal left of Prim 5 stage embryos uninjected and injected as indicated. D: Summary of average length-to-width ratio of the yolk extension of embryos at the Prim 5 stage treated as in (C). E: Summary of the differences in post-anal tail extension of Prim 5 embryos treated similarly to C. F: Percent normal Prim 5 to Prim 15 embryos without gastrulation defects when injected with Sp-MO and/or WT-Rac1 as indicated. Error bars represent the s.e.m for 5 to 118 embryos as indicated on the bars in B, D, and E. Scale bars in A and C = 200 μm. |
|
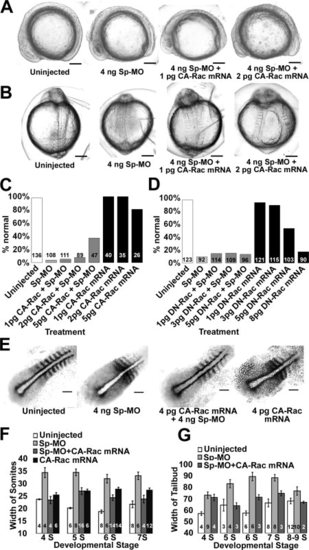
Gastrulation defects caused by p120 catenin δ1 knockdown are partially rescued by co-injection of high concentrations of constitutively active Rac1 GTPase mRNA (CA-Rac1 mRNA), but not with dominant-negative Rac1 mRNA (DN-Rac1 mRNA). A: Side views of embryos, dorsal up, anterior left, at the 11- to 12-somite stages uninjected and injected with Sp-MO and CA-Rac1 mRNA as indicated. B: Dorsal views of the same embryos as in A with anterior up. C: Percent embryos without gastrulation defects at Prim 5 and Prim 15 when injected with Sp-Mo and/or CA-Rac1 mRNA as indicated. D: Percent embryos without gastrulation defects at Prim 5 when injected with Sp-Mo and/or DN-Rac1 mRNA as indicated. E: Micrographs of dissected sibling embryos, dorsal views, anterior to the top right, analyzed with in situ hybridization using a myoD and papc (paraxial protocadherin C) combined probes at the 7-somite stages. Embryos were uninjected or injected as indicated. F: Summary of the average differences in width of the second-to-last somite of embryos treated and probed as in E. Somite numbers are indicated below the bars. G: Summary of the average difference in width of the paraxial mesoderm at the conditions in E and measurements as indicated in Figure 4. The numbers of embryos are given on the graph bars. Scale bars (A, B) = 200 μm and (C) = 100 μm. |
|
Semi-quantitative PCR of cDNAs isolated from Oblong, Bud and 2-Somites stages shows an increase in p120ctn δ2 Chr2 (CTNND2b) after cleavage (oblong stage). Each PCR reaction was 20 μl and contained 2 μl of cDNA from the original isolation for quantitative PCR, 3 mM MgCl2, 10 mM each dNTP, 1 μM each primer, 1.25 U MG Quest Gem Taq, and MG Quest buffer. The reactions were run for 40 cycles (50 cycles for postbud stages) of 15 sec 95oC, 30 sec 55oC, and 30 sec 72oC. The loading control is Ef1a, translational elongation factor 1a; p120ctn δ1 is CTNND1; ARVCF is Armadillo Repeat gene deleted in Velo-Cardio-Facial syndrome; p120ctn δ2 Chr2 is delta catenin or specifically CTNND2b; p120ctn δ2 Chr24 is CTNND2a. The primers for the PCR are given in Supp. Fig. S1: Methods and dot underlined and in bold in Supp. Table S1. Figure S1D shows the change in relative mRNA levels over time relative to the Ef1a loading control determined by the rolling ball method. The amounts at all stages are not as accurate as the quantitative PCR because the amounts were taken a single time point and not the number of cycles required to reach half maximum or the inflection point. EXPRESSION / LABELING:
|
|
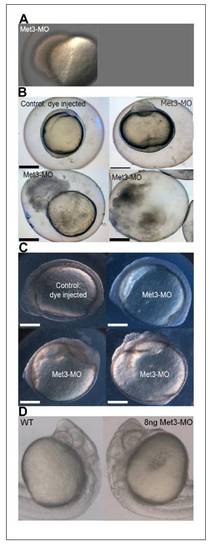
Injection of antisense morpholino RNAs to the Met-3 start site of p120 catenin δ1 disrupts adhesion and causes cells to dissociate from the embryos giving only 12% survival to the 12-somite stage. (A) Injection of 8 -12 ng antisense MO to the 3rd AUG start site of p120 catenin δ1 (Met3-MO) causes a loss of adhesion and embryos disassociate from the yolk cell at 50%-epiboly. Animal pole towards top left. (B) Some embryos injected with 2 ng Met-3-MO imaged at the 1-somite stage slowly disintegrate as cells progressively de-adhere from the dorsal aspect of the embryos. Top-left embryo-dorsal left, anterior up; top-right embryo-ventral view, anterior up. (C) Other embryos injected with 2 ng Met-3-MO reaching the 7- to 9-somite stages have less distinct somite borders than uninjected controls and progressively loose dorsal cells. Top embryos: side views dorsal up, anterior left; Bottom embryos: ventral views, anterior up and toward center. (D) Magnification of Prim 6 stage embryos in side views show defects in the brain when treated with antisense Met-3-MO (8 ng). Scale bars in (B) and (C) are 200 μm. PHENOTYPE:
|