- Title
-
Crosstalk between Fgf and Wnt signaling in the zebrafish tailbud
- Authors
- Stulberg, M.J., Lin, A., Zhao, H., and Holley, S.A.
- Source
- Full text @ Dev. Biol.
|
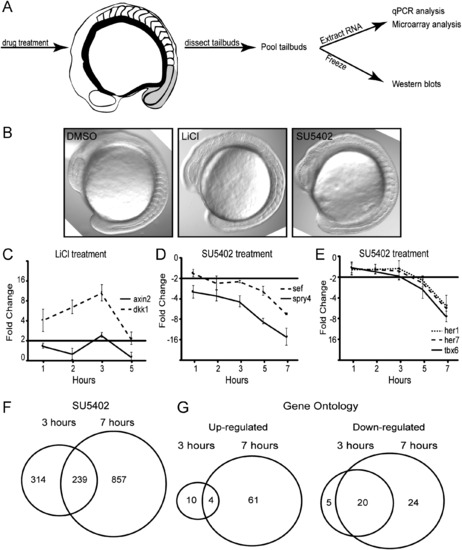
Time course of the transcriptional response in the zebrafish tailbud following activation of Wnt signaling or inhibition of Fgf signaling. (A) Graphical representation of the experimental protocol. (B) DIC images of embryos after 3 h treatment of DMSO, LiCl, and SU5402. (C) Time course of chemical treatment. LiCl activation of Wnt signaling was measured by monitoring transcription of known direct Wnt targets dkk1 and axin2. (D) Time course of SU5402 treatment, an inhibitor of Fgfr1. Fgf signal inhibition was assayed by quantifying transcription of direct Fgf targets sef and spry4. (E) A reduction of presomitic mesoderm cells upon 7 h SU5402 treatment is revealed by reduction in her1, her7, and tbx6 expression. (F) Microarray analysis at the early time point (3 h) and later time point (7 h) suggests that many secondary changes in gene expression are observed at the later time point due to alterations in cell fate and/or cell survival. The data were processed via Agilent′s Feature Extraction software version 10.7.3.1. Samples were normalized using the percentile shift algorithm. No baseline transformation was performed. A 2-fold change cutoff was used to define regulated transcripts. (G) Gene Ontology classification based on Biological Processes Fat indicates that genes representing many more processes are differentially expressed at the later time point than the early, suggesting a loss of cell fates. Fat clustering is a version of Gene Ontologies (GO) that filters out the broadest GO terms so that they do not overshadow more specific GO terms. Fat contrasts to Slim, which provides a broader overview of the ontology content. Error bars are SEM, n=3. |

In situ hybridization of dkk1 following LiCl and SU5402 treatment and qPCR validation of selected microarray data. (A) In situ hybridization of dkk1 following LiCl, SU5402, and LiCl/SU5402 treatment. Embryos were stained in parallel for the same duration and images were processed identically. The spatial pattern of dkk1 does not appear to expand, but levels increase following treatment with LiCl, SU5402, and a combination of LiCl and SU5402. Posterior is down. n=4 experiments, 15 embryos per treatment per experiment. (B) We used direct transcriptional targets of the Wnt and Fgf pathways, along with genes involved in somitogenesis, to validate the microarray data. No change in transcription was observed via microarray or qPCR for genes involved in somitogenesis (her1, her7, hes6) following LiCl treatment. Changes identified via microarray in axin2, dkk1, notum1a, and spred3 expression following LiCl treatment were confirmed by qPCR. Inhibition of Fgf signaling using SU5402 resulted in a change in dkk1, notum1a, fgf8, her1, her7, hes6, sef, sprouty4, dusp4, pea3, spred3, tbx6, and tbx24 expression detected via microarray, and these changes were confirmed by qPCR (n = 4). Error bars are SEM. Statistical comparisons were made using the Student′s unpaired t test. *pd0.06, **pd0.01, ***pd0.001. |
|
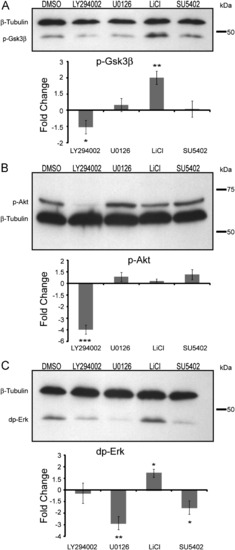
Quantification of Gsk3β, Akt and Erk phosphorylation in the tailbud following perturbation of Fgfr1, Mapk, PI3k and Gsk3β signaling. (A) Western blot analysis of pGsk3β after treatment with LiCl, SU5402, U0126 (Mapk inhibitor), and LY294002 (PI3k inhibitor), reveals levels are significantly down-regulated in LY294002 (n=11) and up-regulated in LiCl (n=9) treatments. (B) pAkt is significantly down-regulated by LY294002 (n=10) and (C) dpErk is down-regulated by U0126 (n=4), SU5402 (n=5), and is up-regulated by LiCl (n=5). Phospho-protein levels normalized to β-Tubulin levels. Statistical comparisons were made using the Student′s unpaired t test. *pd0.05, **pd0.01, ***pd0.001. Error bars are SEM. |
|
In situ hybridization for selected genes following LiCl and SU5402 treatment. We assayed the expression of axin2, her1, hes6, msgn1, notum1a, sef, tbx6, and tbx24. Generally, there was no obvious change in the spatial expression domain, except for the occasional extra her1 stripe due to a shifting of the wavefront. This result had been previously reported in mice expressing a constitutively active beta-catenin in the tailbud (Aulehla et al., 2008; Dunty et al., 2008). In some cases the relative levels of expression are evident, consistent with our microarray and qPCR data. For example, msgn1 and hes6 appear slightly weaker in SU5402 treated embryos. notum1a expression is elevated in discrete domains in the tailbud and head regions following LiCl treatment and elevated specifically in the tailbud following SU5402 treatment. sef expression levels appear slightly elevated after LiCl treatment and are reduced after treatment with SU5402. |
Reprinted from Developmental Biology, 369(2), Stulberg, M.J., Lin, A., Zhao, H., and Holley, S.A., Crosstalk between Fgf and Wnt signaling in the zebrafish tailbud, 298-307, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.