- Title
-
In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains
- Authors
- Moro, E., Ízhan, G., Mongera, A., Beis, D., Wierzbicki, C., Young, R.M., Bournele, D., Domenichini, A., Valdivia, L.E., Lum, L., Chen, C., Amatruda, J.F., Tiso, N., Weidinger, G., and Argenton, F.
- Source
- Full text @ Dev. Biol.
|
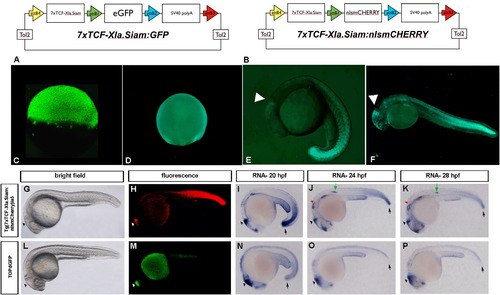
Generation of the TCFsiam lines and comparison with the Tg(TOP:GFP)w25 fish. (A, B) Schematic rapresentations of the Tol-2 based vectors used to generate the Tg(7xTCF-Xla.Siam:GFP)ia4 line (vector shown in A) and the Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 (vector shown in B). (C and D) Strong fluorescence is detectable in Tg(7xTCF-Xla.Siam:GFP)ia4 early staged embryos. Lateral views with anterior to the left are shown in C-P. At dome (C) and 80% epiboly (D) Tg(7xTCF-Xla.Siam:GFP)ia4 transgene expression is evident in all blastodermal cells. (E) During somitogenesis (19 hpf), eGFP is detected in the midbrain-hindbrain boundary (MHB) and in somites with a gradient distribution pattern. (F) 24-hpf embryo. Strong reporter expression is seen in the tail, while the gradient distribution is more clearly visible along the trunk. Positive cells are visible also in the fronto-nasal ectoderm and midbrain-hindbrain boundary (arrowhead). (G?P) Comparison of Wnt reporter activity under epifluorescence and by in situ hybridization in different embryonic stages reveals a stronger and broader reporter activity in Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 embryos than in Tg(TOP:GFP)w25 embryos. (H and M) Reporter fluorescence in Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 embryos is detectable anteriorly in the brain (arrowhead) and posteriorly in tail mesoderm, while in Tg(TOP:GFP)w25 embryos reporter activity is evident only in the brain. (I and N) Reporter transcription in both transgenic lines is detectable in the brain, while Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 additionally display a much stronger activity in the posterior mesoderm at 20 hpf (arrows). (J,K and O,P) After 24 hpf, Wnt reporter transcription is additionally detectable in otic vesicles (red arrow in J and K) and lateral line primordium (green arrow in J and K) in Tg(7xTCF-Xla.Siam:nlsmCherry)ia5, but not in Tg(TOP:GFP)w25 embryos. I-K and N-P are 20, 24 and 28 hpf embryos stained for mCherry and TOPdGFP RNAs, respectively. G and L are brightfield images of the same embryos represented in H and M, respectively. |
|
TCFSiam lines represent bona fide Wnt/β-catenin reporters. (A?F) In situ hybridization with an antisense mCherry probe on Tg(7xTCF-Xla.Siam:nlsmCherry)ia5/ Tg(hsp70l:dkk1-GFP)w32 embryos. In embryos heat-shocked at 50% epiboly, 3 somite-stage and 24 hpf, mCherry RNA is detected at 85% epiboly, 10 somite-stage and 27 hpf, respectively. (A?C) Heat-shocked Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 single transgenic control embryos stained for mCherry RNA. (A) During late gastrula, reporter mRNA is expressed in a broad marginal domain. (B) During somitogenesis, Wnt/β-catenin activity is detectable in the midbrain-hindbrain boundary, and the posterior neuroectoderm and mesoderm. (C) During organogenesis, the reporter is expressed in multiple domains of the CNS, the lateral line primordium, the epidermis of the yolk extension and weakly in posterior tail. (D?F) In heterozygous Tg(7xTCF-Xla.Siam:nlsmCherry)ia5/Tg(hsp70l:dkk1-GFP)w32, mCherry signal is strongly reduced in all expression domains. (G?L) In situ hybridization with an antisense mCherry probe on Tg(hsp70l:wnt8a-GFP)w34/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 embryos. (G?I) Heat-shocked Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 single transgenic control embryos stained for mCherry RNA. Note that the signal intensities in A-C and G-I are different due to variable duration of staining. (J?L) In heterozygous Tg(hsp70l:wnt8a-GFP) w34/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5, a much stronger signal is detected. (M?P) Reporter activity in 55 hpf Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 in apchu745 background (N and P). Wild type siblings (sib) are shown in M and O. Immunostaining for zn8 antibody is visible in blue. Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 expression is upregulated ectopically in the heart, brain region and lateral region (arrows) according to a constitutive activation of the Wnt signaling pathway in apc mutants. (Q,R) Treatment for 36 h with 5 ÁM Alsterpaullone (R) increases Wnt reporter activity in the pharyngeal arches of 3 dpf Tg(7xTCF-Xla.Siam:GFP)ia4 larvae. Parallel treatment with control DMSO (Q) does not alter reporter activity. (S,T) Administration of 0.1 M LiCl for 36 h increases GFP fluorescence in the otic vesicle and pharyngeal arches (white arrowheads) of 3 dpf Tg(7xTCF-Xla.Siam:GFP)ia4 (T) when compared to age-matched transgenic larvae treated with DMSO (S). (U,V). Incubation of transgenic embryos in BIO from 24 hpf to 48 hpf, induces a robust tissue specific activation of the reporter (arrows in V). Notice that BIO treated animals resemble apc phenotype in lateral line, with an hypertrophic, stalled and strongly GFP expressing primordium, while in 2% DMSO control embryos the secondary lateral line primordium is already migrating (asterisk in U). (W,Y) Treatment of 24 hpf Tg(7xTCF-Xla.Siam:GFP)ia4 embryos with 10 ÁM IWR-1 for 24 h (Y) reduced reporter fluorescence particularly at the heart level (white arrow) when compared with DMSO treated embryos (W). ga: gill arches; h:heart, ov:otic vesicle. |
|
Transgene expression in TCFsiam transgenics is highly regulated by lef1. All images (A?D) show a representative 48 hpf larva. Expression analysis of the Tg(7xTCF-Xla.Siam:GFP)ia4 reporter in lef1/sinf U767 homozygous mutants (B) is significantly reduced in several tissues, when compared to age matched Tg(7xTCF-Xla.Siam:GFP)ia4 fish (A). In tcf7l1am881/m881 (hdl) (C) and tcf7l2zf55/zf55 (D) no significant differences can be observed in respect to A. Anterior is shown to the left. Hb: habenula; hy:hypothalamus; t.tectum; pf: pectoral fin; jc: jaw cartilages; MHB: midbrain-hindbrain boundary; h: heart. |
|
TCFsiam expression domains resemble characteristic regions of Wnt/β-catenin signaling activity. (A?H) are reconstructed confocal projections images. (I?L) are images taken with conventional fluorescent microscope. In A?G a comparison between Tg(7xTCF-Xla.Siam:GFP)ia4 and Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 (small inset) show an identical pattern of expression. (A-H) 48-hpf larva. (A) Tg(7xTCF-Xla.Siam:GFP)ia4 is highly expressed in the brain, particularly in the MHB. More caudally, strong Tg(7xTCF-Xla.Siam:GFP)ia4 expression is seen in the spinal cord (B), in the cristae of the otic vesicle (C), in the somitic mesoderm (D), in fin buds (E) and eye(F). (G?H) Posterior lateral line neusromasts actively express the Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 transgene. Confirmation of the neuromasts identity of the cell cluster seen in H (arrowheads) was obtained by Tg(-8.0cldnb:lynEGFP)zf106 transgene coexpression in Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 expressing cells of the neuromasts primordium at 36 hpf of Tg(7xTCF-Xla.Siam:nlsmCherry)ia5/Tg(-8.0cldnb:lynEGFP)zf106 double transgenics. (I) Longitudinal paraffin section of a representative 30 dpf Tg(7xTCF-Xla.Siam:GFP)ia4 larva showing GFP-expressing cells in the retinal pigmented epithelium (rpe). Counterstaining was performed with the nuclear dye DAPI. (J) Representative image of the branchial arch of a one year old Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish demonstrating transgene expression in the tip of gill rackers (gr) and in gill filaments. In the small inset, a magnification of a single gill raker with its fluorescently labeled tip is depicted. (K) Ventral view of a representative forebrain from a one year old Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish, showing strong fluorescence in the hypothalamic region and optic commissure together with labeled endothelial cells of the vascular newtork. (L) Magnification of a representative olphactory bulb labeled in a representative one year old Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish. MHB: midbrain-hindbrain boundary; e: eye; fb: fin bud; e:eye; gf: gill filaments; gr:gill rackers; lln:lateral line neuromasts; ob: olphactory bulb; och: optic commissure; ov: otic vesicle. poc: post-optic commissure; rpe: retinal pigmented epithelium. |
|
Expression analysis of the TCFsiam transgene in wild-type AV canal between 48 and 96 hpf. (A, B, D, E) Confocal images of the AV canal from Tg(myl7:EGFP)twu34/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish (A, D, E) and from Tg(kdrl::EGFP)s843/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 double transgenics (B,C,G,H,I). (F,I) are cross section from the AV canal of Tg(Tie2:EGFP)s849/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 (F) and Tg(kdrl:EGFP)s843/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish (I). (C) Cross section of the bulbus arteriosus (BA) of a 96 hpf Tg(kdrl:EGFP) s843/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish. Green fluorescence has been pseudocolored in blue for Tg(myl7:EGFP) twu34 in D, E, for Tg(kdrl::EGFP) s843 in C,G,H,I and for Tg(Tie2:EGFP) s849 in F. (A?I). Immunostaining with zn8 (blue in A, pseudocolored green in C?I). (A) Few atrial myocardial cells adjacent to the atrioventricular canal (arrowhead) express the Tg(7xTCF-Xla.Siam:nlsmCherry)ia5. (B) Endocardial cells forming the multilayered structure of cardiac valves at 72 hpf express the transgene. (C) Endocardial cells at the boundary between the ventricle and bulbus arteriosus express the transgene (arrowheads). (D, E, G, H, I) round shaped cells at the AV canal adjacent to the cuboidal epithelial cells express the transgene. (D?I) Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 positive cells downregulate alcam/zn8 at the AV boundary and the AV marker Tg(Tie2:EGFP) s849 (F). A:atrium, V:ventricle; AV: atrio-ventricular. |
|
TCFsiam transgene expression in the CNS endothelial compartment during larval, juvenile and adult stages. (A) Confocal image of the trunk vessels in a representative 72 hpf Tg(fli1a:EGFP)y1/ Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish. Few endothelial cells are positive for the transgene (arrowheads in A). (B) Confocal image of the CNS vessels in a 5 dpf representative Tg(fli1a:EGFP)y1/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish. Several endothelial fli1a:EGP expressing cells are positive for the Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 transgene (highlighted by light blue arrows). (C) Confocal Z-stack projection of a 5 dpf Tg(7xTCF-Xla.Siam:GFP)ia4, showing the targeted labeling of the cerebral vascular network. (D) Magnification of a CNS blood vessel of a representative 5 dpf Tg(fli1a:EGFP)y1/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 larva. A double positive endothelial cell is marked by a arrowhead. (E,F) Confocal Z-stack projection of a representative 7 dpf Tg(kdrl:EGFP)s843/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5, showing a perfect localization of the TCFsiam signal in the CNS endothelial compartment. In F a higher magnification of (E) is shown. (G) Confocal Z-stack projection in the midbrain area of a representative 14 dpf Tg(fli1a:EGFP)y1/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish. (H,I) Confocal Z-stack projection of a representative 20 dpf Tg(fli1a:EGFP)y1/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish in the midbrain area (H) and in the forebrain area (I), showing strong labeling of the brain vessels in the transgenic reporter fish. (J) Confocal projection a 30 dpf fish brain showing localized expression of the Tg(7xTCF-Xla.Siam:GFP)ia4 transgene in the vascular compartment of the brain. A small inset illustrates the area analyzed by confocal microscopy in the living fish. (K, L, M) Confocal Z-stack projection of a representative 30 dpf (K) and 60 dpf (M) Tg(fli1a:EGFP)y1/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 and 30 dpf (L) Tg(7xTCF-Xla.Siam:GFP)ia4/Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish showing a persistent expression of the TCFsiam transgene in the brain vascular network. (N) Confocal acquisition of a whole fixed brain from a one year old fish demonstrating long-term expression of the Tg(7xTCF-Xla.Siam:GFP)ia4 transgene in CNS vascular network. |
|
Persistent TCFsiam expression allows fine tracking of neural crest-derived cells. (A) sox10:mRFP expressing cells in otic vesicle of a representative 36 hpf double transgenic larva Tg(sox10:mRFP)vu234/(7xTCF-Xla.Siam:GFP)ia4 coexpress the Tg(7xTCF-Xla.Siam:GFP)ia4 transgene. (B,C) Neural crest-derived cells in the pharyngeal arches coexpress the transgene Tg(7xTCF-Xla.Siam:GFP)ia4 at a high level at 36 hpf. (D) At 72 hpf, Tg(sox10:mRFP)vu234/Tg(7xTCF-Xla.Siam:GFP)ia4 coexpressing cells are dorsal root ganglia (DRG) scattered along the trunk. (E) Montage of a time lapse movie, showing neural crest-derived cells dynamics in the midbrain of a representative 56 hpf Tg(sox10:mRFP)vu234/Tg(7xTCF-Xla.Siam:GFP)ia4 larva. One to two small white arrows in the bottom left indicate migration and neural crest derived cell division along a brain vessel. Note at the 13th frame a small inset showing a doublet of sox10 positive cells coexpressing the TCFsiam transgene. Each frame was recorded every 30 min. (F) Sequential images of a time lapse movie in a representative 5 dpf Tg(sox10:mRFP)vu234/Tg(7xTCF-Xla.Siam:GFP)ia4 larva showing the migration of double positive cells across the somitic mesoderm. Copositive migrating cells are labeled by a white arrow. Each frame was recorded every 1 h. |
|
Wnt/β-catenin signaling is detectable in regenerating tail fins of Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish. (Left column) Bright field live images of unamputated and regenerating fins. (Middle column) mCherry fluorescence in live unamputated and regenerating fins. (Right column) mCherry expression detected by RNA in situ hybridization in unamputated and regenerating fins. (A?C and D?F) Wnt/β-catenin signaling is not detectable in unamputated and newly amputated fish fins, respectively. (G?I) β-catenin activity is detected by strong mCherry RNA expression at 24 hpa while mCherry fluorescence is not visible yet. (J?L) mCherry expression is detectable by both RNA and fluorescence at 48 hpa. (M?O) Wnt/β-catenin signaling is robustly evident by both mCherry RNA and fluorescence at 72 hpa. |
|
|
|
|
|
|
|
|
|
|
|
|
Reprinted from Developmental Biology, 366(2), Moro, E., Ízhan, G., Mongera, A., Beis, D., Wierzbicki, C., Young, R.M., Bournele, D., Domenichini, A., Valdivia, L.E., Lum, L., Chen, C., Amatruda, J.F., Tiso, N., Weidinger, G., and Argenton, F., In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains, 327-340, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.