- Title
-
Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- Authors
- Slagle, C.E., Aoki, T., and Burdine, R.D.
- Source
- Full text @ PLoS Genet.
|
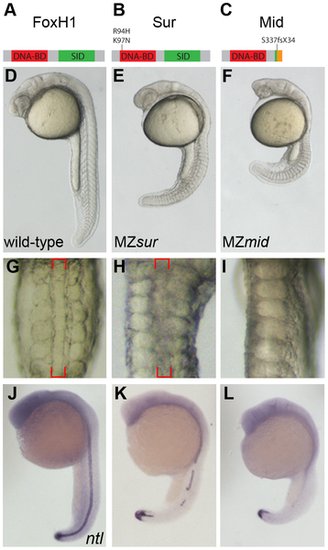
MZmid mutants lack notochords. (A–C) Protein diagrams of FoxH1 alleles. (D–F) 24 hour post-fertilization (hpf) images of live wild-type and maternal-zygotic FoxH1 mutant embryos. (G–I) Dorsal zooms (2.5× relative magnification) of embryos in D–F at the level of the yolk extension. Red brackets mark the notochord; MZmid mutants lack this structure and exhibit midline-fused somites. (J–L) RNA in situ hybridization for ntl expression in 24 hpf wild-type and maternal-zygotic FoxH1 mutants. |
|
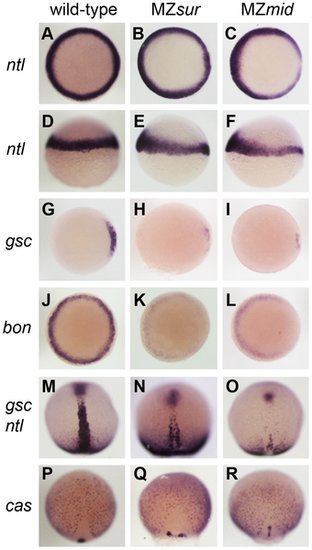
Nodal-dependent tissue specification is differentially disrupted in FoxH1 mutants. (A–L) Mesendoderm marker expression in wild-type and maternal-zygotic FoxH1 mutants at 40–50% epiboly. Dorsal is to the right; A–C, G–L are animal views, D–F are lateral views. Note the dorsal reduction of ntl expression in MZsur (E) and MZmid (F). (M–R) Axial mesoderm and endoderm marker expression at 90% epiboly, viewed dorsally with anterior up. EXPRESSION / LABELING:
PHENOTYPE:
|
|
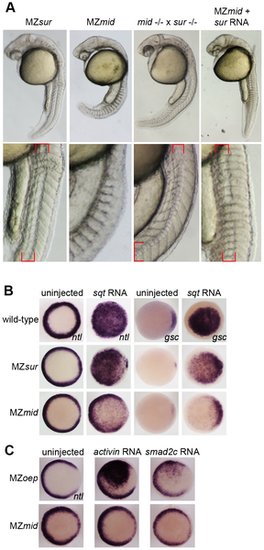
sur retains more Nodal transduction capability than mid. (A) Genetic interactions between the sur and mid alleles, demonstrating the ability of sur to partially rescue the loss of notochord caused by mid. Lower panels are enlargements showing the structures of the notochords at 24 hpf; red brackets indicate notochord domains. (B) Nodal overexpression in maternal-zygotic FoxH1 mutants. 50 pg sqt RNA was injected into wild-type, MZsur, and MZmid embryos at the one-cell stage, and embryos were assayed for Nodal target gene expression at 30–40% epiboly. Note the greater ability of MZsur embryos to respond to ectopic sqt compared to the MZmid response. (C) The mid mutation perturbs activin-like signaling. MZoep and MZmid embryos were injected with RNA encoding either a Xenopus activin homologue (2.5 pg) or an activated form of mouse Smad2 (100 pg). Responses were assayed by observing ntl expression at 30–40% epiboly. EXPRESSION / LABELING:
PHENOTYPE:
|
|
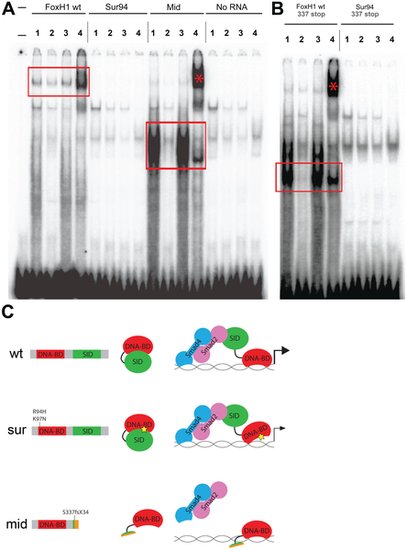
DNA–binding activities of FoxH1 mutants. (A) Electrophoretic mobility shift assays using a FoxH1 binding site probe derived from the zebrafish gsc promoter and in vitro-translated epitope-tagged full-length proteins. FoxH1 and Mid protein bind the probe in a protein- and sequence-specific manner (red boxes and asterisk), while Sur protein shows no binding activity. Upper lane labels indicate the RNA translated for use in each binding reaction; individual lane numbers denote additions to the basic binding reactions (1: no additions; 2: 100-fold excess unlabeled competitor probe; 3: 100-fold excess mutated unlabeled competitor probe; 4: anti-HA antibody). (B) EMSAs using truncated FoxH1 and Sur proteins lacking the C-terminal SID. Truncated wild-type protein specifically binds the probe (red box), while truncated Sur protein does not. Lane markings are as described in (A) above. (C) Model for DNA-binding activities of wild-type and mutant FoxH1 proteins. Wild-type protein binds weakly to its recognition sites alone, but can bind strongly upon loss of its C-terminus, suggesting that Smad interaction may “open up” the conformation of the wild-type protein and allow for strong binding upon pathway activation. Sur protein is impaired in its DNA-binding ability, but may be weakly/transiently tethered to its recognition sequences by activated Smads or other unknown factors. Mid protein cannot interact with Smads and so cannot transduce Nodal signals, but can bind strongly to FoxH1 recognition sites. |
|
Inhibiting Nodal signals in MZmid mutants. Wild-type (A), uninjected MZmid (B), and injected MZmid (C–E) embryos compared to a complete loss of Nodal signaling in MZoep (F) mutants. Inhibiting bon function (3 ng bonMO) in an MZmid background further impairs anterior development but does not affect tail development (C). Blocking eomes function (15–20 pg eomes-enR mRNA; D) resembles both a loss of the Nodal ligands cyc and sqt (8 ng each cycMO+sqtMO; E) and MZoep (F), indicating that Nodal signaling is occurring in MZmid mutants and is mediated by Eomes. PHENOTYPE:
|
|
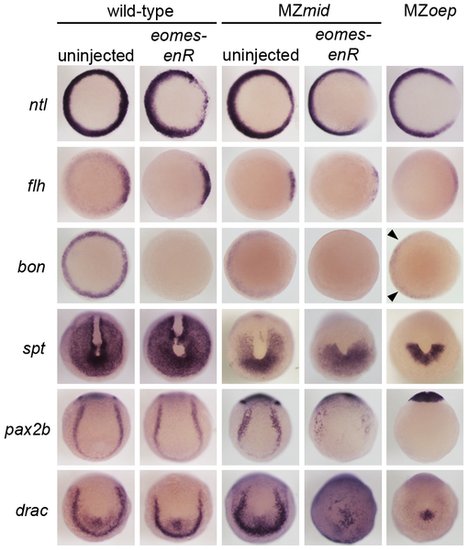
Eomes inhibition enhances the Nodal signaling and mesendoderm deficiencies of MZmid. Markers for various populations of mesendoderm derivatives were analyzed in wild-type and MZmid embryos in which Eomes function was inhibited. Eomes inhibition in wild-type embryos has a minimal effect on expression of most markers, whereas in an MZmid background it has significant effects on mesendoderm-derived tissues, approaching MZoep levels for most markers. Arrowheads indicate Nodal-independent expression of bon in MZoep mutants. EXPRESSION / LABELING:
|
|
Defects caused by overexpression of FoxH1, sur, and mid mRNA. Representative images of common phenotypes observed upon FoxH1, sur, and mid overexpression in wild-type embryos. (A) Embryo injected with 100 pg mid mRNA exhibiting a wild-type appearance at 24 hpf. (B) Embryo injected with 100 pg mid mRNA exhibiting a wavy notochord. (C) Embryo injected with 50 pg FoxH1 mRNA exhibiting a loss of eyes and head structures, and a morphologically irregular notochord (also see panel F). (D) Dorsal anterior view of an embryo injected with 50 pg sur mRNA exhibiting eyes of unequal sizes. (E) Dorsal anterior view of an embryo injected with 50 pg FoxH1 mRNA exhibiting a single unilateral eye. (F) Enlarged portion of embryo in panel C at the level of the yolk extension. Arrows indicate the abnormal notochord. |
|
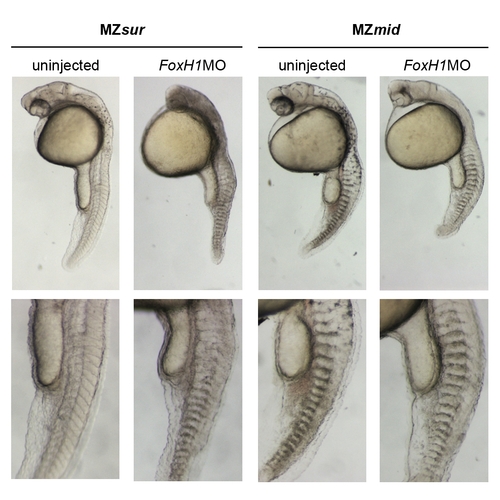
Effects of FoxH1 knockdown on MZsur and MZmid mutants. MZsur and MZmid embryos were injected with 4 ng FoxH1MO to assess the effect of inhibiting production of mutant FoxH1 proteins on development. A majority of injected MZsur embryos lack notochords (115/187 without notochords); injection into MZmid mutants never rescues notochord formation (0/182 with notochords). 15/182 injected MZmid mutants had slight defects including delayed development and nonspecific necrosis which may be injection artifacts. 4/182 displayed midline bifurcations. In 5/182 embryos, injection of FoxH1MO into MZmid caused splitting of the normally fused eye field into two eyes. This effect is most likely related to the published role of the Foxh1 DNA-binding domain in inhibiting the mixl1 promoter via recruitment of Gsc protein in mouse _ [41] (see Discussion for more details). PHENOTYPE:
|
|
Nodal ligand expression in MZmid mutants Pre-gastrulation expression of cyc and sqt were analyzed in uninjected and eomes-enR-injected MZmid embryos. Expression resembles that in wild-type embryos, though at lower levels, and is unaffected by Eomes inhibition. |
|
Functions of ecoptic constitutively active and endogenous Eomes in notochord development. (A) Injection of 25 pg eomes-VP16 frequently causes rescue of notochord formation in MZmid mutants. (B) Injection of eomes-enR does not inhibit notochord formation in wild-type embryos. Arrowheads indicate notochords in injected embryos. |

Unillustrated author statements PHENOTYPE:
|