- Title
-
Retinoic Acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration
- Authors
- Kikuchi, K., Holdway, J.E., Major, R.J., Blum, N., Dahn, R.D., Begemann, G., and Poss, K.D.
- Source
- Full text @ Dev. Cell
|
Resection of the Ventricular Apex Stimulates Immediate, Organ-Wide Morphological and Molecular Changes in Endocardium (A–D) Transmission electron microscope (TEM) analyses of endocardium in uninjured (A) and injured ventricles (B–D). Arrowheads, endocardial nuclei; arrows, endocardial cell bodies; M, cardiac muscle; asterisk, red blood cell. Scale bar represents 2 μm. (E–K) raldh2 expression assessed by in situ hybridization (E–J) and Raldh2 immunostaining (K) in uninjured (E) and injured (F–K) ventricles. Brackets in (I–K), injury site. Arrows in (J and K), epicardial cells. Scale bar represents 100 μm (E–Q). (L–N) Confocal images of altered endocardial cell shape, and enhanced flk1 driven DsRed2 fluorescence in the injury site (L, brackets) in a 7 dpa cmlc2:EGFP; flk1:DsRed2 double transgenic ventricle. Arrowheads, endocardial nuclei; arrows, endocardial lining of myofiber. An antibody against DsRed was used. DAPI (42-6-Diamidino-2-phenylindole) stains nuclei. (O–Q) Sections of 7 dpa fli1:EGFP (O), hand2:EGFP (P), or gata5:EGFP (Q) transgenic ventricles. Brackets, injury sites. Arrowheads in inset, Raldh2+/EGFP+ endocardial nuclei with rounded morpholog |
|
Induction of raldh2 Expression in Various Injury Models (A and B) Stab injuries into the ventricular apex assessed for raldh2 induction (A) and fli1:EGFP expression (B) at 7 days poststab (dps). Arrows, needle entry site. Scale bar represents 100 μm. (C) Confocal image of Raldh2 immunofluorescence in fli1:EGFP+ endocardial cells with rounded morphology at the injury site (arrowheads). Scale bar represents 20 μm. (D–H) raldh2 induction after intraperitoneal LPS or vehicle (PBS) injection. Scale bar represents 100 μm (D–L). (I–L) Endocardial raldh2 (J), hand2 (K), and gata5 (L) expression surrounding spontaneous infarcts (asterisks) within cultured ventricular explants. Dead cardiac tissue was identifiable by the absence of cell nuclei (I). |
|
Transgenic Inhibition of RA Signaling Blocks Cardiomyocyte Proliferation during Regeneration (A) Assessment of Mef2+PCNA+ cells (arrows) in wild-type (wt) and hsp70:dn-zrar transgenic fish at 7 dpa, after a single heat-shock at 6 dpa. Maximum projection images of 10 μm Z stacks are shown. Insets, high-magnification images of the rectangle. Arrowheads, proliferating epicardial cells; brackets, injury site. Scale bar represents 100 μm (A and C). (B) Quantification of CM proliferation in wt and hsp70:dn-zrar transgenic fish at 7 dpa. Data are mean ± standard error of mean (SEM) from 6 animals each (3097 wt and 2482 transgenic CMs analyzed). *p < 3 × 10-5, Student′s t test. (C) Assessment of Mef2/PCNA double-positive cells (arrows) in wt and hsp70:cyp26a1 transgenic fish at 7 dpa, after a single heat-shock at 6 dpa. Maximum projection images of 10 μm Z stacks are shown. (D) Quantification of CM proliferation in wt and hsp70:cyp26a1 transgenic fish at 7 dpa. Data are mean ± SEM from 4 wt and 6 transgenic animals (3888 wt and 4760 transgenic CMs analyzed). *p < 2 × 10-4, Student′s t test. |
|
Cardiac Injury Responses in Polypterus, Mouse, and Zebrafish (A–D) raldh2 (p. raldh2) expression by in situ hybridization in uninjured (A) and injured (B–D) polypterus ventricles. Arrowheads in (A), pigment cells. Brackets in (B–D), injury site. Insets in (B–D), lateral ventricular wall including epicardium (ep). Arrows in (D), p. raldh2-expressing epicardial cells. Scale bar represents 100 μm (A–H). (E–G) Assessment of Mef2+PCNA+ cells (arrows) in uninjured (E) and injured (F and G) polypterus ventricles. Brackets in (F), injury site. Insets, high-zoom images of the rectangle. Arrowhead in (G), entry site of glass needle. Arrows in (F and G), proliferating CMs. (H) In situ hybridization of p. raldh2 after stab injury (arrowhead). (I–L) Raldh2 (m. Raldh2) expression by in situ hybridization at various time points post-ligation (pl). Insets in (J–L), high-zoom images of the rectangle. ep, epicardium; en, endocardium; LVL, left ventricular lumen. Scale bar represents 200 μm. (M) Summary of injury responses observed in polypterus, zebrafish, and mouse hear |
|
(A) heg in situ hybridization shows weak or no detectable expression in the uninjured ventricle. Scale bar = 100 μm (A-C). (B) heg is induced by 3 hours post-amputation (hpa) in endocardial cells throughout the ventricle (and atrium, not shown). (C) raldh2 in situ hybridization at 1 hpa, indicating induction in many atrial endocardial cells, but fewer ventricular endocardial cells. The injured ventricular apex is marked by an asterisk. |
|
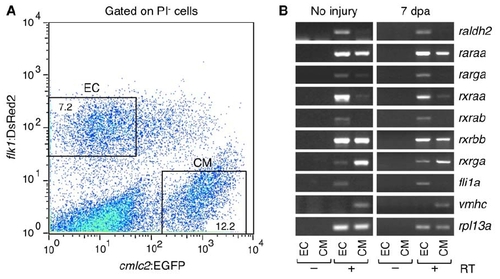
Expression Analyses of RA Signaling Components (A) Endocardial cells (EC) and cardiomyocytes (CM) were purified by FACS from dissociated uninjured or 7 dpa cmlc2:EGFP; flk1:DsRed2 ventricles. A representative plot from 7 dpa ventricles is shown. Cells stained by propidium iodide (PI) are excluded as dead cells. Percentages of the total PI-negative cell population that were gated are shown. (B) Expression of RA signaling components was examined by RT-PCR in purified endocardial cells (EC) and cardiomyocytes (CM). Endocardial (fli1a) and cardiomyocyte markers (vmhc) were used to confirm the specificity of cell sorting. ribosomal protein L13a (rpl13a), was used as a control. The amplified products were not detectable without reverse transcriptase (RT), confirming that bands are amplified from cDNAs of the target genes but not from genomic DNA contaminants. raldh2 transcripts detected in samples of uninjured ventricles are likely accentuated by the severe trauma of ventricular cell extraction and dissociation. |
|
Effects of dn-zrar Induction during Embryogenesis and Regeneration (A-D) A single heat-shock at 37°C for 30 minutes was delivered to hsp70:dn-zrar or wild-type (wt) embryos at 24 hours post-fertilization (hpf) as described previously (Lee et al., 2005). At 72 hpf, cardiac edema (B, arrowheads) and pectoral fin reductions (D, arrowheads) were observed in hsp70:dn-zrar embryos, but not in wt (A, C, arrowheads). Cardiac edema was observed in all embryos with pectoral fin defects. (E, F) Cardiomyocyte expression and nuclear localization of the dn-zrar inhibitory product (green) in sections of uninjured (E) and regenerating (F) ventricles. Most or all cardiomyocytes show strong nuclear dn-zrar expression one day after a heat-shock. Scale bars = 100 μm in (E, F, H, and I). (G) A second line of hsp70:dn-zrar was analyzed at 7 dpa for cardiomyocyte (CM) proliferation, after a single heat-shock at 6 dpa. wt, clutchmate control. Data are mean ± SEM of 4 animals analyzed (2364 wt and 3038 transgenic cardiomyocytes analyzed). *p < 3 x 10-8, Student′s t-test. (H, I) Apoptosis was examined by TUNEL assay and quantified in wt and hsp70:dn-zrar ventricles one day after a single heat-shock. TUNEL signals were quantified at the apex for uninjured ventricles, and at the apical injury site at 2 dpa and 7 dpa. TUNELpositive CMs were rare in uninjured animals, and appeared at similar frequencies at 2 dpa in wt and hsp70:dn-zrar injuries. Unlike 7 dpa wt ventricles, some 7 dpa transgenic ventricles displayed several TUNEL-positive cells within the myocardium (arrows in bracketed area of (I)), but overall there were no significant differences at the tested timepoints. Apoptotic cells were detected in the clot (asterisk) in both wt and transgenic ventricles. The dotted lines indicate approximate borders of the clot. Data are mean ± SEM of 3-9 animals analyzed. |
Reprinted from Developmental Cell, 20(3), Kikuchi, K., Holdway, J.E., Major, R.J., Blum, N., Dahn, R.D., Begemann, G., and Poss, K.D., Retinoic Acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration, 397-404, Copyright (2011) with permission from Elsevier. Full text @ Dev. Cell