- Title
-
Spatio-temporal regulation of Wnt and retinoic acid signaling by tbx16/spadetail during zebrafish mesoderm differentiation
- Authors
- Mueller, R.L., Huang, C., and Ho, R.K.
- Source
- Full text @ BMC Genomics
|
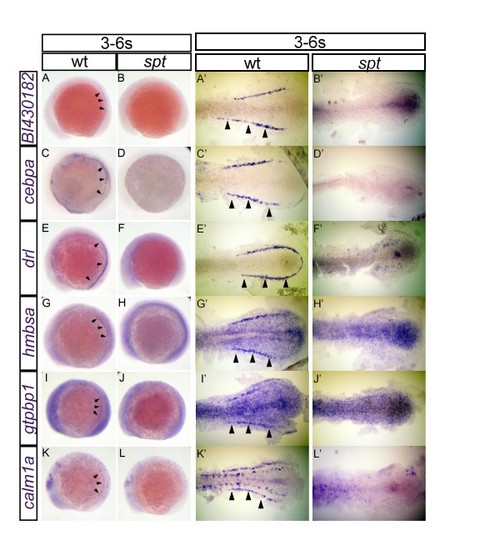
Expression of validated candidate genes from the 4/5-somite microarray. Changes in the expression of BI430182 (A, B, A′, B′), cebpa (C, D, C′, D′), drl (E, F, E′, F′), hmbsa (G, H, G′, H′), gtpbp1 (I, J, I′, J′) and calm1a (K, L, K′, L′) were visualized using in situ hybridization in wild-type (A, A′, C, C′, E, E′, G, G′, I, I′, K and K′) and spt embryos (B, B′, D, D′, F, F′, H, H′, J, J′, L and L′). All embryos are oriented with posterior to the right. Whole-mount embryos (A-L) are shown in lateral view while de-yolked and flat-mounted embryos (A′-L′) are shown in dorsal view. Flat-mounted embryos (A′-L′) show only the posterior half of the embryos for high-magnification view. All embryos were fixed at the 3 to 6-somite stage. Arrowheads point to specific intermediate mesoderm expression domain EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression of validated candidate genes from the 21-somite microarray. Changes in the expression of aldh1a2 (A, B), rcan3 (C, D), bmp3 (E, F), hsp90a.1 (G, H), unc45b (I, J) and fgfrl1a (K, L) were visualized using whole mount in situ hybridization in wild-type embryos (A, C, E, G, I and K) and spt embryos (B, D, F, H, J and L). All embryos are shown in lateral view with anterior to the left. All embryos were fixed at the 21-somite stage. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

Unillustrated author statements |