- Title
-
Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development
- Authors
- Znosko, W.A., Yu, S., Thomas, K., Molina, G.A., Li, C., Tsang, W., Dawid, I.B., Moon, A.M., and Tsang, M.
- Source
- Full text @ Dev. Biol.
|
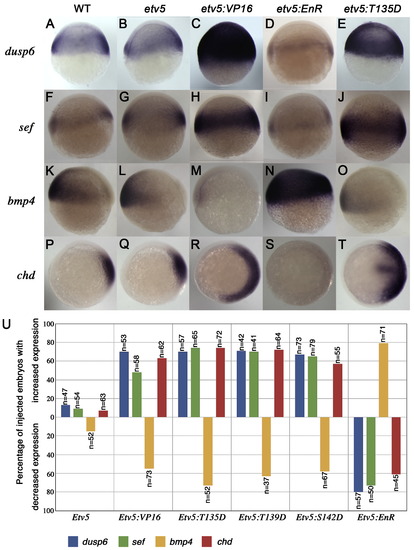
Etv5 functions as a positive effector in FGF signaling. (A–O) Lateral views and (P–T) animal views at shield stage. In situ probes and constructs injected are indicated on the left and above, respectively. Expression of downstream targets of FGF signaling, dusp6 and sef, are unaltered in etv5-injected embryos (B, G), but were induced by etv5:VP16 mRNA injections (C, H). Ectopic expression of dominant negative (etv5:EnR) blocks dusp6 and sef expression (D, I). Injection of etv5:T135D increased dusp6 and sef expression (compare A, F to E, J). Expression of bmp4 and chordin was unchanged in etv5-injected embryos (L, Q). A dorsalized phenotype was noted in etv5:VP16-injected embryos (M, R) and a ventralized phenotype upon etv5:EnR mRNA injection (N, S). Injection of etv5:T135D mRNA evoked dorsalized phenotypes (O, T). (U) Frequency of phenotypes elicited by microinjection of mRNAs indicated. |
|
The Pea3 ETS factors perform redundant functions and are required for mid–hindbrain boundary (MHB) formation. (A–I) Lateral views of 28hpf embryos. Injection of antisense morpholinos targeting pea3, erm and etv5 (3EtsMO) resulted in MHB defects (B) as compared to the Control scrambled morpholino (A). 3EtsMO-induced MHB defects can be rescued by co-injection of HAetv5 or HApea3 mRNA (D–F) but not by GFP mRNA (C). Expression of dusp6 is diminished in 3EtsMO-injected embryos (H). Co-injection of HAetv5 and HApea3 rescued dusp6 expression (I). Red arrowhead marks loss of MHB in MO-injected embryos. |
|
Pea3 ETS factors are required for target gene responses to FGF signaling. (A–P) Lateral views of embryos injected with MOs indicated above and assayed with probes indicated on the left. Expression of FGF target genes, dusp6 and sef, were unchanged in etv5MO-injected embryos (B, F). In contrast, the double or triple knockdown of Pea3 ETS genes elicited a drastic reduction of dusp6 and sef (C, D, G, H). Knockdown of Pea3 ETS genes resulted in MHB defect as measured by her5 (I–L) and pax2a (M–P) expression. Individual knock-down had minimal effects, while 3EtsMO injections resulted in dramatic reduction of her5 (L) and pax2a (P). (Q) Graph depicting the phenotypic effects of etv5MO, 2EtsMO and 3EtsMO on expression FGF target genes and MHB markers. Red arrowhead marks loss of MHB gene expression. |
|
Pea3 ETS factors are required to maintain cardiac progenitors. (A–X) Dorsal views at 10-somite stage. In situ probe listed on left and MO above. (A–D) nkx2.5 expression was reduced as multiple Pea3 factors were knocked down (D). This was similar to fgf8MO knock-down and in ace mutants (E–F). (G–J; M–P) gata4 expression (G–J) and hand2 expression (M–P) were also reduced after Pea3 depletion (J, P). A similar phenotype was observed in fgf8MO knock-downs and in ace mutants (K, L;Q, R). (S–V) Dorsal view of scl expression indicated expansion of endothelial lineages in EtsMO injections (T–V) as indicated by arrowheads that mark the caudal limit in uninjected embryos (S). Similar results were noted in fgf8MO and ace embryos suggesting that FGF signaling is required to maintain cardiac progenitors and to limit endothelial lineages to the rostral ALPM (W, X). (Y) Graph providing quantitative data for MO experiments. |
|
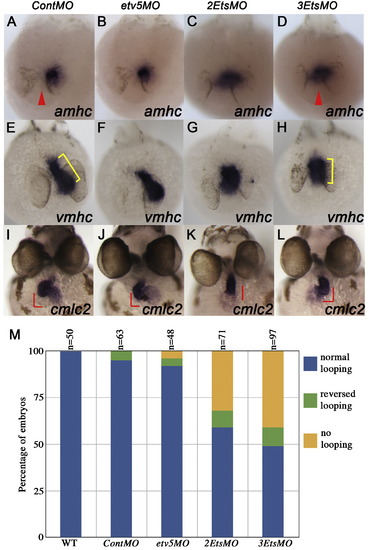
Pea3 ETS factors are required for heart development. (A–L) In situ hybridization showing cardiac gene expression at 24hpf (A–H) and 48hpf (I–L). Knock-down of multiple Pea3 ETS factors altered amhc (D; red arrowhead indicates midline of embryo) and vmhc expression (H; cell population and spatial arrangement in yellow bracket). Cardiac looping was severely affected in 3EtsMO-injected embryos as shown by expression of cmlc2 (K, L). (M) Quantification of effects on heart looping in MO-injected embryos as exemplified in I; normal looping, K; no looping, L; reverse looping. |
|
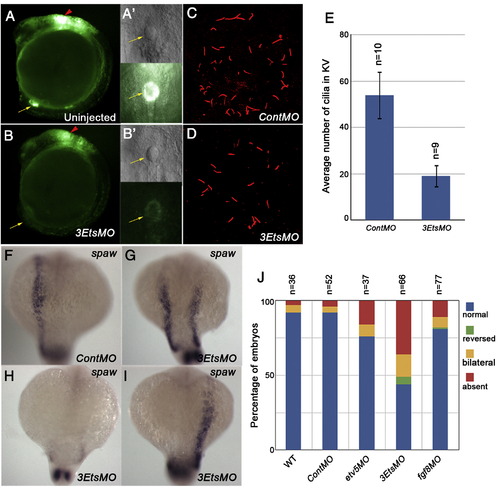
Pea3 ETS factors are required for ciliogenesis in the Kupffer′s vesicle and subsequent L/R patterning. (A, B) Tg(dusp6:EGFP)pt6 embryos injected with 3EtsMO show grossly normal formation of Kupffer′s vesicle, even though FGF signaling was suppressed (B, B′). (C, D) Cilia number in the KV was reduced in 3EtsMO-injected embryos at the 12-somite stage. (E) Graph showing average number of cilia in MO-injected embryos. Expression of spaw in 3EtsMO-injected embryos reveals L/R defects at 18-somite stage (F–I). (J) Graph depicting L/R defects in Pea3 ETS and Fgf8 depleted embryos. |
|
Etv5-ETS domain binds to the Dusp6 promoter. (A) Biotin labeled PCR product. (B) Biotin labeled mutant Pea3B Dusp6 promoter fragment. (C) Pea3B 18 bp fragment used in competition assays. Underlined sequence represent the putative Pea3B binding site (D) EMSA using 60 bp Dusp6 promoter biotin fragment and GST-Etv5:ETS protein. The Etv5-ETS domain bound to the 60 bp Dusp6 promoter containing the Pea3B site (lane 2). This binding was competed with the non-biotin labeled 60 bp Dusp6 promoter (lanes 3–7) and the Pea3B 18 bp fragment (lanes 8–12). A random oligonucleotide sequence did not compete this binding (lanes 13–17). Binding of Etv5-ETS to the Pea3B mutant Dusp6 promoter was not observed (lanes 18–19). |
|
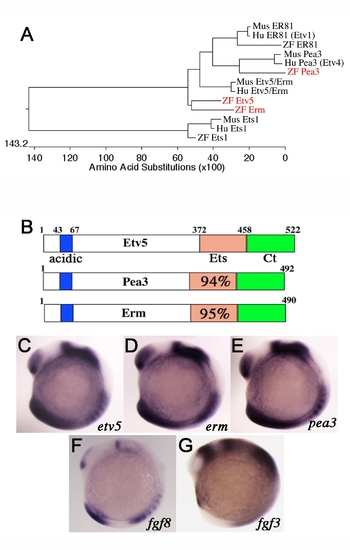
Comparative expression of Pea3 ETS transcription factors during zebrafish development. (A) Phylogenetic alignment of selected zebrafish (red), mouse and human PEA3 ETS factors. (B) Diagram of structural features of Pea3 ETS proteins showing the conserved acidic region (acidic), ETS DNA binding domain (Ets), and carboxyl region (Ct). Percent identities between Etv5 ETS domain to Pea3 and Erm are listed. (C–G) Lateral views at 10-somite stage. In situ probes indicated on the bottom right corner. Pea3 ETS transcription factors have overlapping expression patterns and are similar to that of fgf3 and fgf8 ligands. |
|
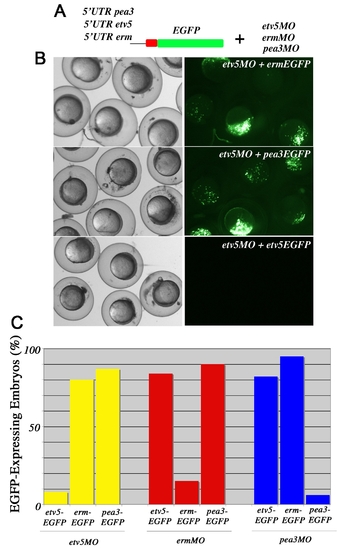
Specificity of Pea3 ETS MO. (A) Experimental design to determine the specificity of each of the Pea3 ETS MOs. (B) Brightfield live images of shield stage embryos on the left and EGFP expression on the right. The construct and MO injected are indicated in the upper right corner. Embryos co-injected with the 5′UTR erm:EGFP DNA and etv5MO at 2–4 cell stages or with 5′UTR pea3:EGFP DNA and etv5MO at 2–4 cell stages expressed EGFP. In contrast, over 90% of embryos microinjected with 5′UTR etv5:EGFP DNA and etv5MO did not show GFP expression, indicating the efficiency of etv5MO. (C) A graphical representation of all possible combinations of ETS MO and Ets-eGFP, supporting that each MO is specific for the knock-down of only one Pea3 ETS family member. |
|
Pea3 ETS factors are required for foxj1a expression. (A) Expression of foxj1a in contMO injected embryos at 90% epiboly stage. (B) In 3EtsMO-injected embryos, foxj1a is significantly reduced. |
Reprinted from Developmental Biology, 342(1), Znosko, W.A., Yu, S., Thomas, K., Molina, G.A., Li, C., Tsang, W., Dawid, I.B., Moon, A.M., and Tsang, M., Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development, 11-25, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.