- Title
-
A systematic approach to identify functional motifs within vertebrate developmental enhancers
- Authors
- Li, Q., Ritter, D., Yang, N., Dong, Z., Li, H., Chuang, J.H., and Guo, S.
- Source
- Full text @ Dev. Biol.
|
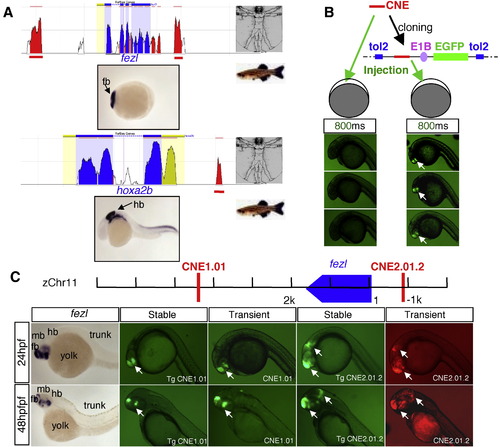
In vivo enhancer detection in zebrafish. (A) A schematic showing identification of CNEs (red peaks) near anterior brain-expressed or posterior brain-expressed genes through comparative bioinformatics analysis using the ECR browser (Ovcharenko et al., 2004) between human and zebrafish. (B) Schematic diagram showing the enhancer detection construct composed of cloning sites for CNEs, E1B basal promoter, and reporter in Tol2 transposon vector, and comparison of two enhancer detection methods. With 800 ms exposure time, the GFP signal is robust in embryos injected with cloned CNE (right), whereas the signal is below detection in embryos injected with PCR product of CNE (left). (C) Comparison of reporter signals in transient versus stable transgenic embryos. CNE1.01 and CNE2.01.2 are identified near the fezl gene. Similar activity is observed between transient (CNE1.01, CNE2.01.2) and stable (Tg CNE1.01, Tg CNE 2.01.2) transgenic embryos. |
|
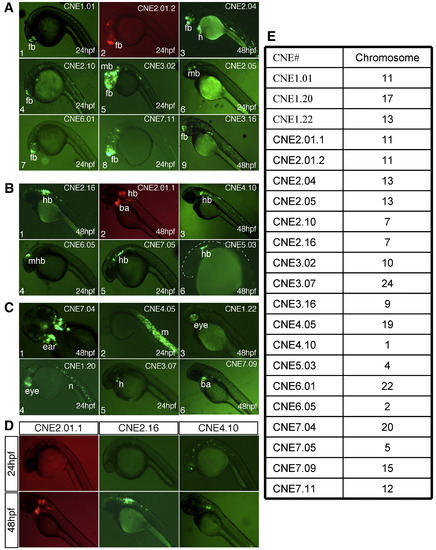
Analysis of pattern-associated CNEs reveals spatial and temporal enhancers. (A) CNEs with anterior brain enhancer activity. (1) CNE 1.01 shows specific enhancer activity in sub-regions of the forebrain (fb). (2) CNE2.01.2 drive reporter (mCherry) in the fb and midbrain (mb). (3) CNE2.04 drives GFP specifically in the fb, heart (h), and few muscles cells in the trunk region. (4) CNE2.10 drives reporter expression primarily in the telencephalon. (5) CNE3.02 shows strong enhancer activity in the anterior brain. (6) CNE2.05 drives reporter expression in sub-regions of the ventral mb. (7) CNE6.01 drives reporter expression primarily in the telencephalon. (8) CNE 7.11 has enhancer activity in fb and mb, extending into anterior hindbrain (hb). (9) CNE 3.16 drives strong GFP expression in sub-regions of the telencephalon. (B) CNEs with posterior brain enhancer activity. (1) CNE2.16 drives reporter expression in the anterior hb. (2) CNE2.01.1 is able to drive reporter expression in the anterior hb and branchial arch (ba) regions. (3), (4) CNE4.10 and CNE 7.05 drive reporter expression in the ventral hb. (5) CNE6.05 drives reporter expression in the midhindbrain boundary (mhb). (6) CNE5.03 drives reporter expression in subsets of anterior hb cells. (C) CNEs with other tissue-specific activity. (1) CNE7.04 is an ear enhancer. (2) CNE4.05 is able to drive reporter expression in the trunk muscle (m). (3) CNE1.22 is an eye enhancer. (4) CNE1.20 drives reporter expression in eyes, hb, and notochord (n). (5) CNE3.07 is a heart enhancer. (6) CNE7.09 drives reporter expression in ba regions. (D) CNEs with temporal specific activity. CNE2.01.1 and CNE2.16 have no detectable enhancer activity at 24 hpf, but have distinct enhancer specificity at 48 hpf. CNE4.10 is able to drive reporter expression at 24 hpf with a non-specific pattern, and drive reporter expression specifically in sub-regions of hb at 48 hpf. (E) Table showing the chromosomal location (based on Zv7) of the CNEs shown. |
|
High-resolution analysis of anterior brain enhancer activity. Triple labeling and confocal analysis of CNE reporter expression in zebrafish embryos. (A–B) CNE1.01 shows restricted activity in dorsal posterior telencephalon and distinct cell clusters in the hypothalamus. (C–D) CNE2.01.4 shows distinct activity in anterior and posterior telencephalon but not in medial telencpehalon, as well as sub-regions of the prethalamus. (E–F) CNE2.04 shows activity in posterior telencephalon, prethalamus, thalamus, and pre-tectal regions. (G–H). Schematic diagrams showing the anterior brain expression domains of the three CNEs shown. Abbreviations: Hy, hypothalamus; Pret, pretectum; Pt, prethalamus; Tel, telencephalon; Th, thalamus. |
|
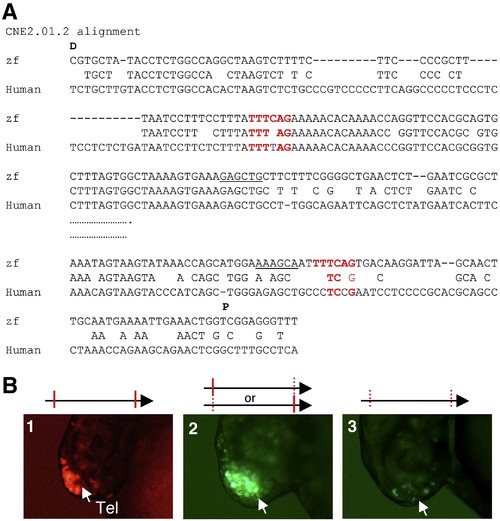
Mutating the motif TTTCAG impairs the forebrain enhancer activity of CNE2.01.2. (A) Alignment of zebrafish and human CNE 2.01.2. Abbreviations: D, distal to the transcription start site; P, proximal to the transcription start site. (B) Functional validation of de novo predicted motifs in the CNE2.01.2. Reporter activity of (1) The CNE containing the wildtype copies of the motif TTTCAG, or mutations in the following motif: (2) either the distal or the proximal copy of the motif; (3) Mutating both copies of the motif. |
|
Mutating the motifs AATGAA, AATGGA, GAGCGG, and GAGGGG impairs the forebrain enhancer activity of CNE1.01. (A) Alignment of zebrafish and human CNE 1.01. Abbreviations: D, distal to the transcription start site; P, proximal to the transcription start site. (B) Functional validation of de novo predicted motifs in the CNE 1.01. Reporter activity of (1) The CNE containing wildtype copies of the predicted motifs, or with the following motifs mutated: (2) the distal copy of the motif AATGAA; (3) the proximial copy of the motif AATGAA; (4) the motif GAGCGG; (5) the motif AATGAA; (6) and (7) the motif GAGGGG. |
|
Selected images showing brain region-specific gene expression. |
|
CNEs as versatile tools to driving gene expression in desired tissues. |
Reprinted from Developmental Biology, 337(2), Li, Q., Ritter, D., Yang, N., Dong, Z., Li, H., Chuang, J.H., and Guo, S., A systematic approach to identify functional motifs within vertebrate developmental enhancers, 484-495, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.