- Title
-
Zebrafish gbx1 refines the midbrain-hindbrain boundary border and mediates the Wnt8 posteriorization signal
- Authors
- Rhinn, M., Lun, K., Ahrendt, R., Geffarth, M., and Brand, M.
- Source
- Full text @ Neural Dev.
|
Overexpression of gbx1 induces posterior cell fate. (A) Wild-type (WT) embryo at 24 h and (B) after injection of 200 pg gbx1 mRNA; anterior brain structures are severely reduced. The ear is indicated by an arrowhead. (C, D) Staining of the forming axon tracts with an anti-acetylated tubulin antibody at 24 h; dorsal views, with anterior to the top. In the gbx1-injected embryo the hindbrain is severely enlarged compared to WT (arrowheads). (E, F) Duplications of ear structures are frequently observed (arrows). (G-I′) Series of embryosinjected with different doses of gbx1 mRNA (200 and 500 pg), analyzed at the tailbud stage after in situ hybridization with otx2 (blue) and krox20 (red). The otx2 domain progressively disappears and the krox20 domains shift to more anterior regions. (J) Dose-dependent gbx1 overexpression phenotypes. Higher concentrations (>500 pg) did not increase the observed phenotype. (A-D, K-M) Lateral views; (E-H, K′-M′) dorsal views. |
|
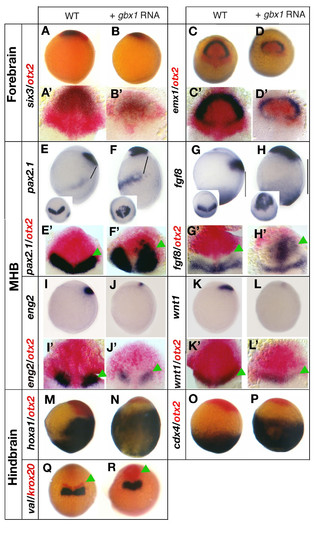
gbx1 overexpression affects midbrain, midbrain-hindbrain boundary (MHB) and hindbrain. (A, A′) Control embryo stained for six3 (blue) and otx2 (red);. (B, B′) gbx1-injected embryo. The near absence of otx2-positive cells posteriorly to the six3-expressing domain indicates a loss of the midbrain territory in the gbx1-injected embryos. (C, C′) Control embryo stained for emx1 (blue) and otx2 (red). (D, D′) gbx1-injected embryo. The emx1 domain corresponding to forebrain remains robust, whereas the otx2 domain corresponding to midbrain is lost. (E) Wild-type (WT) embryo after in situ hybridization (ISH) with pax2.1 and (E′) with pax2.1 and otx2 (red). (F, F′) gbx1-injected embryos; pax2.1 is expressed ectopically in the anterior of the embryo. The MHB is shifted anteriorly as indicated by green arrowheads. Also, the distance between the posterior pax2.1 domain and the anterior domain is increased (black bars in (E, F)). (G) WT embryo after ISH with fgf8 and (G′) with fgf8 and otx2 (red). (H, H′) gbx1-injected embryos; fgf8 is expressed ectopically in the anterior of the embryo. The MHB is shifted anteriorly as the distance between this expression domain and the margin in increased (black bars in (G, H)). (I) WT embryo after ISH with eng2 and (I′) with eng2 and otx2 (red). (J) gbx1-injected embryo; eng2 is not expressed ectopically. (J′) Combined ISH with otx2 (red) shows that the remaining domain is located caudally to the reduced otx2 domain. (K) WT embryo after ISH with wnt1 and (K′) with wnt1 and otx2 (red). (L) gbx1-injected embryo;wnt1 is not expressed ectopically. (L′) In all gbx1-injected embryos the MHB is shifted anteriorly as indicated by green arrowheads. (M) WT embryo after ISH with hoxa1 and otx2 (red). (N) gbx1-injected embryo; the hoxa1 expression doamin is enlarged. (O) WT embryo after ISH with cdx4 and otx2 (red). (P) gbx1-injected embryo; the cdx4 expression domain is enlarged. (Q) WT embryo after ISH with val and krox20 (red). (R) gbx1-injected embryo; val expression is not affected: the krox20 expression domain corresponding to rh3 (green arrowhead) is enlarged, whereas rh5 is less affected. (A, B, E-P) Lateral views, anterior to the left; (C, D-Q, R) dorsal views, anterior to the top; (A′-L′) flat-mounted embryos, anterior to the top. EXPRESSION / LABELING:
|
|
gbx1 acts as a mediator of the Wnt8 hindbrain posteriorizing signal. (A) Control embryo at 24 h, (B) gbx1-injected embryo (200 pg), (C) wnt8-injected embryo (400 pg). gbx1 overexpression mimics wnt8 gain-of-function; the embryos are truncated anteriorly. The red arrowhead indicates the position of the ear. (D-G′) Animal pole views at 60% epiboly after ISH with otx2. (D) Control embryo, (E, E′) gbx1-injected embryos (200 pg), (F) embryo injected with wnt8 morpholinos, (G, G′) embryos co-injected with gbx1 mRNA and wnt8 morpholinos. Expansion of the otx2 domain in the wnt8 morphant is rescued by co-injection with gbx1 mRNA. (H-K′) Dorsal views at the tailbud (tb) stage after ISH with otx2 (blue) and krox20 (red). (H) Control embryo, (I, I′) gbx1-injected embryos, (J) embryo injected with wnt8 morpholinos, (K, K′) embryos co-injected with gbx1 mRNA and wnt8 morpholinos. Loss of krox20 and posterior expansion of the otx2 domain in the wnt8 morphant is rescued by gbx1 mRNA. |
|
gbx1 induces posterior neural fate via transformation of anterior neural fate and gbx1 and otx2 repress each other cell-autonomously. (A) Schematic drawing of the fluorescein uncaging procedure (for details, see [33]). (B, C) gbx1-injected embryo after uncaging cells in the prospective otx2 domain. (D, E) After 20 h of development the labeled cells come to lie in the anterior of the embryo (arrow). (B, D) Nomarski optics; (C, E) detection of the fluorescent cells (arrow). (F) Experimental procedure for cell transplantations. Donor embryos are generated by co-injecting biotinylated dextran as lineage tracer and gbx1 (300 pg) or otx2 mRNA (300 pg). Cells are taken out of the donor at 40% epiboly and transplanted into a wild-type (WT) host embryo. (G) Chimeric embryo containing cells derived from embryos injected with biotinylated dextran and gbx1mRNA, and stained for otx2 (blue). Unlabeled patch of cells marked by a white arrow. (G′) Close-up of the patch indicated in (G) before biotin staining and (G″) after biotin staining (brown). gbx1 overexpressing cells within the otx2 domain do not express otx2. (H) Chimeric embryos containing cells derived from embryos injected with biotinylated dextran and otx2 mRNA, and stained for gbx1 (blue). Unlabeled patch of cells marked by a black arrow. (H′) Close-up of the patch indicated in (H) before biotin staining and (H″) after biotin staining (brown). otx2 overexpressing cells within the gbx1 domain do not express gbx1. |
|
gbx1 morpholinos affect hindbrain patterning. (A-C′) Embryos at the four somite stage stained for otx2/krox20/myod. (A) Control embryo, (B, B′) embryos injected with 10 ng Mo1, (C, C′) embryos injected with 7 ng Mo2. Injections of two different morpholinos show the same phenotype morphologically and after in situ hybridization analysis. (D-F) Rescue experiments using the krox20 expression domain as a read-out. (D) gbx1-morphant, (E, E′) rescued morphant embryos co-injected with gbx1 mRNA that does not contain the MO binding sequence in its 5′ region. (F) Two combinations were tested: 10 ng of Mo1 with 200 pg or 400 pg of gbx1 mRNA. (G, H) Western blot detection showing down regulation of gbx1-myc mRNA translation in embryos co-injected with gbx1-myc mRNA containing morpholino binding sequences and Mo1 (G) or Mo2 (H). WT, wild type. EXPRESSION / LABELING:
PHENOTYPE:
|
|
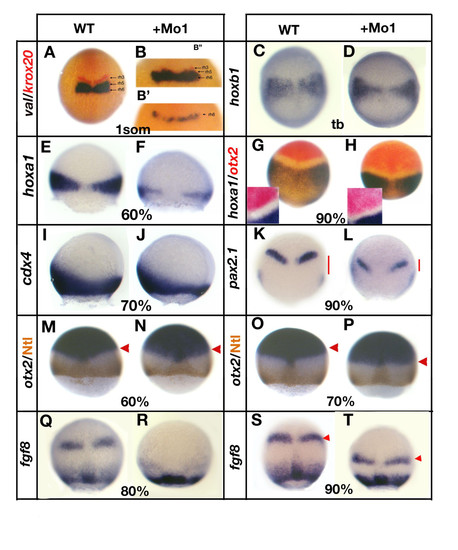
Loss of gbx1 affects hindbrain and midbrain-hindbrain boundary (MHB) markers. (A-H, M-K-T) are dorsal views, (I, J) are lateral views and the developmental stages are indicated (tb, tailbud stage). (A) Control embryo (wild type (WT)) stained with val (blue) and krox20 (red), (B, B′) gbx1-morphants. A residual krox20 expression is seen in (B′) and no expression of krox20 in (B″). For (B′, B″) a higher magnification of the region of interest is shown. (C) Control embryo stained with hoxb1; (D) gbx1-morphant. (E) Control embryo stained with hoxa1; (F) gbx1-morphant. (G) Control embryo stained with hoxa1 and otx2; (H) gbx1-morphant. A higher magnification is shown, illustrating the reduction of size of the gap between the otx2 and hoxa1 domains. (I) Control embryo stained with cdx4; (J) gbx1 morphant. (K) Control embryo stained with pax2.1; (L) gbx1-morphant. The red bars show the distance between the MHB and the lateral plate mesoderm expression domains. This distance is markedly reduced in the gbx1 morphants. (M, O) Control embryos stained with otx2 (blue) and Ntl antibody (brown); (N, P) gbx1 morphants. The otx2 domain is unaffected at 60% epiboly but slightly shifted posteriorly at 70% (red arrowheads). (Q, S) Control embryos stained with fgf8; (R, T) gbx1-morphants. MHB expression is delayed (R) and shifted posteriorly (S, T, red arrowheads) in the morphants. |
|
The otx2/gbx1 interface plays an essential role in midbrain-hindbrain boundary development. (A) Lateral view of a chimeric embryo stained for fgf8. The red arrowhead indicates ectopic fgf8 expression. (B) Lateral view of a chimeric embryo stained for pax2.1. The red arrowhead indicates ectopic pax2.1 expression. (C) Lateral view of a chimeric embryo stained for eng2. The red arrowhead indicates ectopic eng2 expression. (A′, B′, C′) Close-up of the ectopic patch before biotin staining and (A″, B″, C″) after biotin staining (brown). For all the three genes, ectopic expression is induced in gbx1-overexpressing cells within the otx2 domain (red arrowheads). (D) Summary table of the transplanted cells' positions and marker induction. Clones localized in the forebrain never induced ectopic fgf8, pax2.1 or eng2 expression. |
|
Comparison of gbx1 Mo1 and Mo2 induced phenotypes. (A-F) Lateral views, anterior to the top (the tailbud stage) and to the left (24 h). (G-K′) Dorsal views, anterior to the top. (A) Control embryo at the tailbud stage; (B, B′) embryos at the tailbud stage injected with 5 ng and 10 ng Mo1 respectively; (C, C′) embryos at the tailbud stage injected with 5 ng and 10 ng Mo2 respectively. (D) Control embryo at 24 h; (E) 24 h embryo injected with 5 ng Mo1; (F) 24 h embryo injected with 5 ng Mo2. |
|
krox20 expression at somite stage in gbx1 morphants. (A) Control embryo at the seven-somite stage stained with otx2/krox20/myoD. otx2 is not seen in this picture. The most anterior expression seen is the krox20 domain. myoD indicates the number of somites. (B) Embryos at the seven-somite stage injected with 7 ng of Mo1. The most anterior expression seen is the border of the otx2 domain and krox20 is not visible. EXPRESSION / LABELING:
|