- Title
-
A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation
- Authors
- Morley, R.H., Lachani, K., Keefe, D., Gilchrist, M.J., Flicek, P., Smith, J.C., and Wardle, F.C.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
Ntl binds and regulates the expression of genes involved in posterior identity. (A) Diagrams to show fgf and wnt signaling pathways. Components outlined in black were identified as Ntl targets by ChIP-chip. (B–F) Ntl binding in genomic regions around wnt8, fgf24, vent, vox, sp5l, and aldh1a2. Plots show ChIP-enrichment ratio for microarray probes in the genomic regions. Chromosomal position, TSS, intron-exon structure, putative upstream TBSs with constrained character (green lines; see text), and conserved CRMs (red bar; see text) are shown below the graphs. (G–N2) In situ hybridization of ntl mutants compared to wild types for vent, vox, sp5l, and aldh1a2 (posterior views, ventral down). Expression of vent, vox, and sp5l shows downregulation at bud stages (arrows). In addition expression of sp5l is absent from the dorsal forerunner cells in ntl mutant embryos (asterisk; DFCs outlined by dashed line). aldh1a2 expression in the ventral margin is downregulated at gastrula stages. |
|
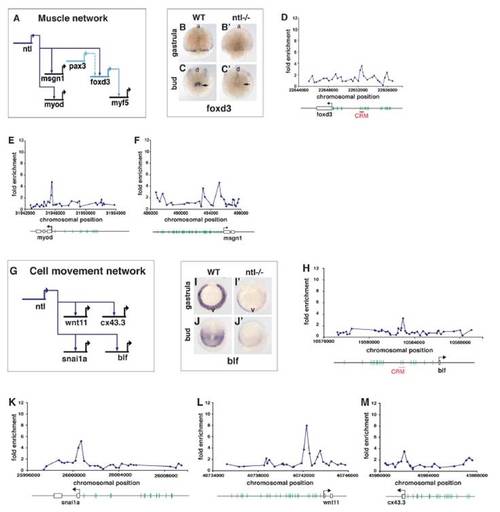
Ntl binds and regulates expression of genes involved in muscle specification and morphogenetic movements. (A) GRN showing Ntl binding and regulation of factors involved in specifying muscle cell fate. Solid lines indicate binding of target promoter and genetic regulation. Dashed line indicates target binding only or genetic regulation without proven target binding. (B–C′) In situ hybridization of ntl mutants compared to wild types at gastrula and bud stages shows downregulation of foxd3 (arrows). (D–F) Ntl binding in genomic regions around foxd3, myod, and msgn1. Plots show ChIP-enrichment ratios for microarray probes in the genomic regions shown. Chromosomal position, TSS, intron-exon structure, putative upstream T-binding sites with constrained character (green lines; see text), and conserved CRM (red bar; see text) are shown below the graphs. (G) GRN showing Ntl binding and regulation of genes involved in morphogenetic movements. (I–J′) In situ hybridization shows downregulation of blf in ntl mutant compared to wild-type embryos at 70% epiboly and bud stage. (H–M) Ntl binding in genomic regions around blf, snai1a, wnt11, and cx43,3. a, anterior; d, dorsal; v, ventral |