- Title
-
The fifth class of Gα proteins
- Authors
- Oka, Y., Saraiva, L.R., Kwan, Y.Y., and Korsching, S.I.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
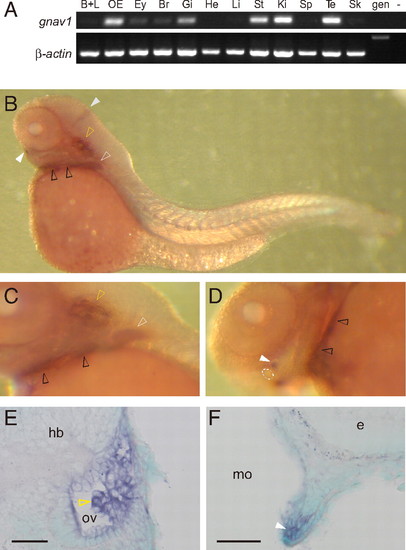
Expression pattern of the zebrafish gnav1 gene. (A) Transcripts for zebrafish gnav1 were detected by RT-PCR using intron-spanning primer pairs. β-Actin was used as a positive control. B+L, barbels and lips; OE, olfactory epithelium; Ey, eye; Br, brain; Gi, gill; He, heart; Li, liver; St, stomach; Ki, kidney; Sp, spleen; Te, testis; Sk, skin; gen, genomic DNA; -, negative control without template DNA. (B–F) Whole-mount in situ hybridization with gnav1 probe. (B and C) Lateral views of the whole larva and posterior part of the head region, respectively. (D) Ventral view of the head region. Dotted circle, mouth. (E and F) Cross-sections after hybridization show expression in the developing inner ear (E) and lower lip (F). Sections were counterstained with methyl green. Dorsal is to the top. White and gray solid arrowheads indicate the cell clusters next to the lower lip and the midbrain–hindbrain boundary, respectively. White, yellow, and black open arrowheads point to labeled cells within pectoral fins, otic vesicle (ov), and branchial arches, respectively. e, eye; hb, hindbrain; mo, mouth cavity. (Scale bars: 50 μm.) EXPRESSION / LABELING:
|
|
Expression pattern of gnav1 in zebrafish larvae: whole-mount in situ hybridization of gnav1 probe (A–C and G–J) and negative control (sense probe, D–F and K–N) with 3 dpf zebrafish larvae. Embryos were grown in 0.0045% of phenylthiourea (Sigma) from 12 h postfertilization until used to prevent pigmentation. Probes were synthesized with T3 RNA polymerase (Roche) by following manufacturer’s instructions. Templates for in vitro transcription were amplified using primers listed in Table S3. Probes were hybridized at 65 °C overnight. Specific probe hybridization was detected with anti-DIG antibody conjugated with alkaline phosphatase (1:5,000; Roche), using Nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate (Roche) as substrates. Stained larvae were observed and photographed with a Nikon SMZ-U binocular and an attached Nikon CoolPix 950 digital camera. Stained larvae were cryo-sectioned (8 μm), counterstained in 0.001% methyl green, mounted with VectaMount (Vector), and documented on a Zeiss Axioplan microscope and an attached AxioCamMRc5. (A and D) Lateral view of whole larvae. (B and E) lateral view around the developing inner ear. (C and F) Ventral view of head region. Dotted circle, mouth. (G–N) Cross-sections of stained larvae at the levels of developing inner ear (G and K), mouth (H and L), pectoral fin (I and M), and branchial arches (J and N). Dorsal is to the top. White and gray arrowheads indicate the cell clusters near the lip and midbrain–hindbrain boundary, respectively. White, yellow, and black arrowheads point to labeled cells within pectoral fin (pf), otic vesicle (ov), and branchial arches (ba), respectively. e, eye; hb, hindbrain; mo, mouth cavity; op, operculum; ph, pharynx; y, yolk. (Scale bars: 50 μm.) |
|
Expression pattern of gnav compared to gna genes of the other 4 classes. Whole-mount in situ hybridization of gnav, gnas, gnat2, gnao1, gna11a, and gna13a probes with 3 dpf zebrafish larvae was performed as described in Fig. S3. All images are lateral views, and anterior is to the left. Note that expression patterns are characteristically different and none from the other 4 classes is similar to that of gnav1. Colored frames enclose genes from the same class (red, Gv; blue, Gs; purple, Gi; green, Gq; yellow, G12). Primers used to clone gna genes are as follows: gnas-fw, 5′-aagactgaggaccagcgaaa-3′; gnas-rv, 5′- gctggacaggctaactggac-3′; gnat2-fw, 5′-ctggtgaagctgccacagta-3′; gnat2-rv, 5′-gcttctctacaagcgccatt-3′; gnao1-fw, 5′-ccagtccaacgctgtctttt-3′; gnao1-rv, 5′- cgctccttgtctccgtactc-3′; gna11a-fw, 5′-cgatcaggttctggtggaat-3′; gna11a-rv, 5′-tgaaaggcgagttggagtct-3′; gna13a-fw, 5′-agaaactgcacatcccttgg-3′; and gna13a-rv, 5′-ttttggctgggcaagtagtc-3′. |