- Title
-
Tcf3 inhibits spinal cord neurogenesis by regulating sox4a expression
- Authors
- Gribble, S.L., Kim, H.S., Bonner, J., Wang, X., and Dorsky, R.I.
- Source
- Full text @ Development
|
Analysis of tcf3a and tcf3b spinal cord expression and effectiveness of morpholinos. (A-F) Cross-sections with the spinal cord outlined in red. tcf3a and tcf3b are co-expressed in the intermediate and ventral spinal cord at 15 hpf (A,D), 18 hpf (B,E) and 24 hpf (C,F). (G,H) RT-PCRs for both tcf3a and tcf3b splice morphants show unspliced products (the asterisk in each gel) at 5 ng/nl for each morpholino. (I,J) tcf3 splice morphant embryos lack eyes, produce minimal brain tissue rostral to the midbrain-hindbrain boundary (MHB) and have a shortened rostral-caudal axis. EXPRESSION / LABELING:
PHENOTYPE:
|
|
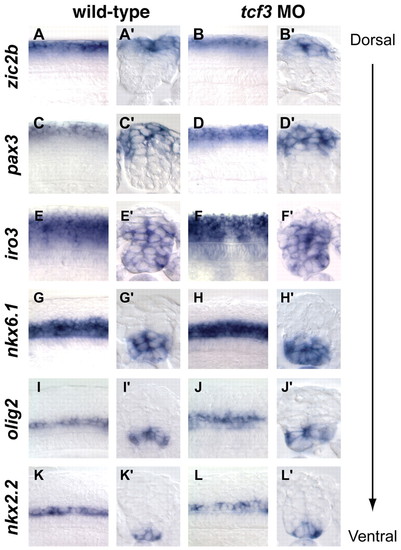
Dorsoventral patterning is unaffected in tcf3 morphants. Whole-mount lateral and cross-section views of 18 hpf wild-type and tcf3 morphant embryos hybridized for zic2b (A-B′), pax3 (C-D′), iro3 (E-F′), nkx6.1 (G-H′), olig2 (I-J′) and nkx2.2 (K-L′). None of these expression domains was unaffected in tcf3 morphant embryos. EXPRESSION / LABELING:
|
|
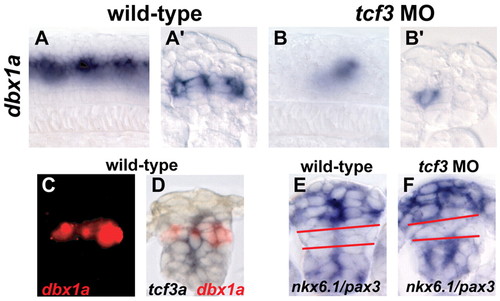
dbx1a expression is absent in tcf3 morphants. (A,A′) Whole-mount lateral and cross-section views of 18 hpf wild-type embryos hybridized for dbx1a mRNA. (B,B′) tcf3 morphant embryos hybridized for dbx1a, showing loss of expression throughout intermediate spinal progenitors. Residual expression is likely to be in postmitotic neurons. (C,D) Double in situ hybridization for dbx1a (red) and tcf3 (blue), showing overlap between the two genes. (E,F) Both wild-type (E) and tcf3 morphants (F) have a similar gap (red lines) between the pax3 (dorsal) and nkx6.1 (ventral) expression domains, suggesting that a general dorsoventral shift in patterning has not occurred. EXPRESSION / LABELING:
|
|
Tcf3 controls the rate of spinal progenitor proliferation. (A-F) The mitotic index (A-C) and the BrdU-labeling index (D-F) of tcf3 morphant (MO) embryos are slightly decreased, yet significantly different (*), as compared with wild-type (WT) embryos. Error bars show s.e.m. and P-values were calculated using unpaired t-tests. (B,C) Whole-mount views of anti-pH3 staining, with the spinal cord demarcated by the black lines. (E,F) Cross-sections, with BrdU staining in green and propidium iodide (PI) counterstaining in red. (G-J) In situ hybridization on cross-sections shows that proliferative cell nuclear antigen (pcna) and cdkn1c (p57) are expressed normally in tcf3 morphants at 18 hpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Spinal progenitors lacking Tcf3 precociously express neuronal markers. (A,B) Cross-sections of tubb5 in situ hybridization at 18 hpf show that tcf3 morphants have ectopic tubb5-positive cells in the medial progenitor domain (arrows). (C,D) Double labeling for HuC/D (green) and BrdU (red) at 24 hpf shows BrdU-positive Hu-positive cells (arrows) in tcf3 morphants, some of which are in the medial domain. (E-N) Lateral whole-mount views of postmitotic neuronal markers at 18 hpf. (E-H) lhx1a and pax2a, which mark multiple classes of spinal interneurons, are drastically reduced in tcf3 morphants. (I-N) evx1+ interneurons, en1b+ interneurons and isl1+ primary motoneurons (asterisks) are also reduced in tcf3 morphants. EXPRESSION / LABELING:
|
|
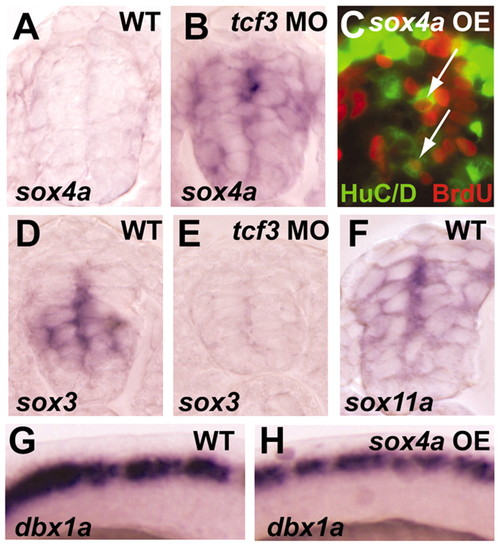
sox4a is repressed by Tcf3 in spinal progenitors. (A,B) sox4a is weakly expressed in the spinal cord of wild-type embryos at 24 hpf and is ectopically expressed throughout the intermediate and ventral spinal cord of tcf3 morphants. (C) Overexpression of sox4a mRNA leads to ectopic HuC/D BrdU double-positive cells in medial progenitors (arrows). (D,E) sox3 is expressed in intermediate and ventral spinal progenitors of wild-type embryos at 24 hpf, and is drastically reduced in tcf3 morphants. (F) sox11a is expressed in both spinal progenitors and postmitotic neurons at 24 hpf. (G,H) sox4a overexpression does not affect the expression of dbx1a in spinal progenitors. |
|
A repressor form of Tcf3 can block neurogenesis upstream of sox4a. (A,B) Heat shock-induced expression of ΔTcf at 18 hpf eliminates expression of sox4a, both alone and in the presence of tcf3 morpholinos. (C,D) ΔTcf expression at 18 hpf completely blocks BrdU uptake at 24 hpf. (E,F) Expression of ΔTcf (green, visualized by GFP fusion) blocks expression of HuC/D (red), both alone and in the presence of tcf3 morpholinos. (G) Simultaneous overexpression of sox4a mRNA and the ΔTcf transgene leads to ectopic Hu-positive cells in the medial spinal cord (arrow). (H) Co-injection of tcf3 and sox4a morpholinos does not lead to ectopic Hu-positive cells in the medial spinal cord. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Tcf3 binds to regulatory regions of sox4a in vivo. (A) A Tcf3a polyclonal antibody detects a specific band that is absent from tcf3 morphants, and detects larger bands in ΔTcf3-expressing embryo lysates. Multiple bands are most likely to be due to rearranged copies of the transgene. (B) ChIP analysis of whole-embryo lysates shows specific immunoprecipitation of two phylogenetically conserved DNA fragments in regulatory regions of sox4a. A third fragment, lacking nearby Tcf sites, does not precipitate. The conservation plot is adapted from the UCSC Genome Browser. ChIP fragments are indicated by red bars and potential Tcf-binding sites in conserved regions are indicated by green ovals |

Unillustrated author statements EXPRESSION / LABELING:
PHENOTYPE:
|