- Title
-
An Fgf8-dependent bistable cell migratory event establishes CNS asymmetry
- Authors
- Regan, J.C., Concha, M.L., Roussigne, M., Russell, C., and Wilson, S.W.
- Source
- Full text @ Neuron
|
The fgf8 Mutant Has a Symmetric Epithalamus (A–D) Dorsal views of confocal images of the epithalamus in wild-type and ace 3 dpf embryos, with anterior to the top. (A and B) 3D reconstructions of pineal/parapineal nuclei and axons [green, Tg(foxD3:GFP)] and neuropil of the habenular nuclei (red, anti-acetylated tubulin; white-edged arrowheads). (C and D) The parapineal-specific marker Tg(ET11:GFP) is present (green), but expressing cells remain at the midline in the ace embryo compared with those of the wild-type embryo at 3 dpf. Brain morphology is visualized using the nuclear marker TOPRO3 (red). A single z-slice is presented for each example. (E and F) Frontal view of parapineal-specific gfi expression in wild-type and ace 3 dpf embryos, with dorsal to the top. Parapineal cells are at the midline in ace (F). (G–J) Dorsal views of habenular lov (G and H) and brn3a (I and J) expression in wild-type and ace embryos at 4 dpf; expression of both markers is reduced in left and right habenulae. EXPRESSION / LABELING:
|
|
Temporally Controlled Abrogation of Fgf Signaling Identifies a Critical Window for Fgf-Dependent Parapineal Migration (A and C–E) Expression of fgf8 in the epithalamus at 24 hpf ([A], dorsal view, TOPRO3 nuclear marker in gray, parapineal primordium highlighted in blue), 28 hpf ([C], lateral, black arrowhead; [D], dorsal, black arrowhead denotes stronger left-sided expression), and 36 hpf ([E], dorsal). Dashed ellipse indicates position of pineal nucleus (D and E). (B) Schematic depicting pineal organ (po), parapineal nucleus (pp, blue), fgf8 expression domains (red), and midline (dashed line), as visualized in (A). (F) fgfR4 expression at 36 hpf (dorsal). Dashed lines indicate position of pineal nucleus (large ellipse), parapineal nucleus (small ellipse), and midline (straight line). (G–J) 3D reconstructions and (G′–J′) single z-slices of dorsal views of the epithalamus in control- (G, G′, I, and I′) and SU5402- (H, H′, J, and J′) treated Tg(foxD3:GFP) 4 dpf embryos, with anterior to the top. Pineal cells have been pseudocolored in blue in single z-slices and dashed lines indicate position of pineal nucleus and midline (G′–J′). SU5402 treatment at 24–28 hpf completely abolished the initial leftward migration of the parapineal to lateral and dorsal positions (H, and H′). Treatment at 36–44 hpf abrogated later translocation of the parapineal to ventral and medial locations relative to the pineal nucleus, and parapineal cells remain at dorso-lateral positions adjacent to the pineal ([J and J′], white arrowheads). L, left; R, right. |
|
Local Provision of Fgf8 Restores Parapineal Migration in ace Embryos and Directs Laterality of Migration in the Absence of a Nodal Signaling Bias (A, E, I, and M) Frontal views of lefty1 expression (as a marker of Nodal signaling) in ace (A), wild-type (E), ntlMO/ace (I), and spawMO/ace (M) embryos at 20 hpf, with dorsal to the top. (B, F, J, and N) dorsal views of live brains showing Fgf8-soaked beads implanted rostrally and to the left (B), right (J and N), and midline (F) of the pineal complex visualized by Tg(foxD3:GFP) expression, in ace (B), wild-type (F), ntlMO/ace (J), and spawMO/ace (N) embryos. Anterior is to the top. (C, C′, D, D′, G, G′, H, H′, K, K′, L, L′, O, O′, P, and P′) 3D reconstructions and single z-slices of dorsal views of the epithalamus in ace (C–D′), wild-type (G–H′), ntlMO/ace 1(K–L′), and spawMO/ace (O–P′) Tg(foxD3:GFP) embryos at 3 dpf implanted with BSA- (C, C′, G, G′, K, K′, O, and O′) or Fgf8- (D, D′, H, H′, L, L′, P, and P′) loaded beads. Pineal/parapineal complex (green) is visualized as before. Pineal cells have been pseudocolored in blue in single z-slices. Anterior is to the top. (Q) Graph representing proportions of embryos with right (green), left (blue), or static (gray) parapineal nuclei after epithalamic implantation of BSA- and Fgf8-loaded beads in ace embryos, ace embryos with modulated Nodal, and spawMO embryos. Results are grouped according to position (left, middle, or right) of bead implantation. EXPRESSION / LABELING:
|
|
Fgf8 is expressed in the epithalamus throughout asymmetric development (A-C) Expression of fgf8 in the epithalamus at 22 ss (A), 48 hpf (B) and 72 hpf (C) (dorsal view). |
|
Bead implants locally up-regulate targets of Fgf8 signalling, but do not affect habenular development or Nodal gene expression (A-D) lov expression at 3dpf is unaffected by implantation of BSA- (A,C) or Fgf8- (B,D) loaded beads at 26 ss in wild-type (WT; A,B) and ace mutant (C,D) embryos. |
|
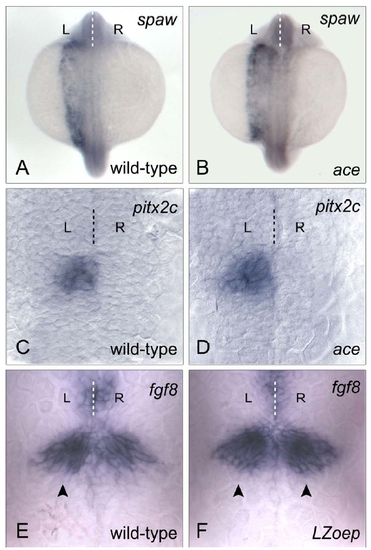
ace mutants have normal axial and epithalamic Nodal signalling; subtle asymmetries in fgf8 expression are Nodal-dependent (A,B) Axial expression of the Nodal ligand encoding southpaw (spaw) gene is unaffected in ace mutant embryos: 97.5 % of ace embryos (B) had left-sided spaw expression compared to 99 % of wild-type embryos (A) at 20 ss. |