- Title
-
Fgfs control homeostatic regeneration in adult zebrafish fins
- Authors
- Wills, A.A., Kidd Iii, A.R., Lepilina, A., and Poss, K.D.
- Source
- Full text @ Development
|
Inhibition of Fgf signaling causes progressive tissue loss from zebrafish fins. (A) Images of wild-type and hsp70:dn-fgfr1 fins at day 0, day 30 and day 60 of heatshock. Wild-type fins were unaffected by daily heatshocks, whereas transgenic fins showed progressive loss of distal fin tissue. Fins shown are representative, and not from the same animal at each timepoint. (B) High-magnification images of distal fin structures after 30 days of heat shock. Many hsp70:dn-fgfr1 rays exhibited severe tissue loss, which was often accompanied by an excess of epidermal tissue (arrowheads). (C) Quantification of fin loss by measurement of centrally located rays (see Materials and methods). hsp70:dn-fgfr1 animals displayed significant reductions in fin length following both 30 and 60 days of heatshock, whereas wild-type clutchmates showed no changes (mean±s.e.m.; Student's t-test, *P<0.001 at days 30 and 60). (D) Dorsal and anal fins of hsp70:dn-fgfr1 zebrafish showed fin atrophy after 60 days of Fgfr blockade. The fins of wild-type clutchmates retained their length and morphology after 60 days of similar heat treatments. PHENOTYPE:
|
|
Fgf receptor inhibition causes pathology at intersegmental joints. (A) Many hsp70:dn-fgfr1 fins exhibited swelling (arrow) or dislocation (arrowhead) of the ray segments at the intersegmental joints (bottom). A representative image of a wild-type fin is provided for comparison (top). (B) Confocal images of whole-mount hsp70:dn-fgfr1 and wild-type fins after 30 days of heatshock, stained with zn3 antibody to visualize scleroblasts. (Left) Example of scleroblast expansion at hsp70:dn-fgfr1 segment joints (bottom) in a case of visible joint pathology (arrowheads). (Middle) Joint hypertrophy was also observed in regions of hsp70:dn-fgfr1 fins without obvious structural damage (arrowheads). (Right) Segmental joints viewed at high magnification, with expansion and disorganization in an hsp70:dn-fgfr1 joint (brackets). (C) Sections of hsp70:dn-fgfr1 and wild-type fins after 30 days of heatshock, stained to visualize scleroblasts with zns5 antibody. There is an expanded zone of rounded scleroblasts (red) surrounding the segment joints of transgenic hemirays. (Left) Regenerates at 4 days post-amputation also have rounded scleroblast morphology. |
|
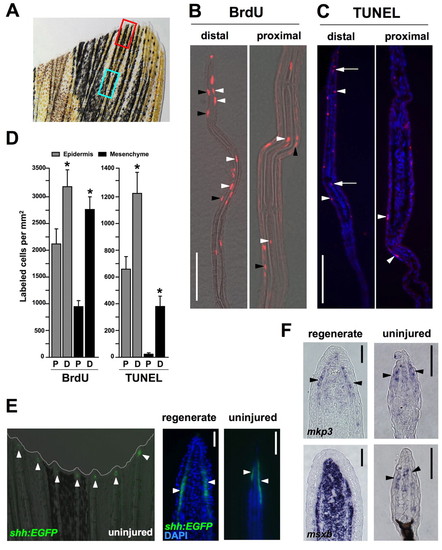
Homeostatic regeneration programs active in zebrafish fins. (A) Ventral lobe of an uninjured caudal fin. The red box indicates areas taken for distal measurements; the blue box is for proximal measures. Each box is 350 μm long, separated by 350 μm, a length chosen because it is the length of a frame at 20x magnification using our imaging equipment. (B) Distal and proximal sections of the caudal fin stained for BrdU incorporation, after a 24-hour labeling period. BrdU-labeled cells (red) are observed in epidermal (black arrowheads) and mesenchymal (white arrowheads) compartments of both distal and proximal regions (distal to top of each image). (C) TUNEL stains of distal and proximal regions, labeling apoptotic cells. TUNEL-positive cells (red) are observed in the epidermal (arrowheads) and mesenchymal (arrows) compartments. Nuclei are labeled with DAPI (blue). (D) Quantification of BrdU and TUNEL labeled cells in epidermal and mesenchymal compartments of proximal (P) and distal (D) fin tissue. (mean±s.e.m.; Student's t-test, *P<0.05). (E) Analysis of a shh:EGFP transgenic reporter strain. (Left) Whole-mount detection of EGFP fluorescence at the distal tips of each ray (arrowheads) of an uninjured shh:EGFP transgenic zebrafish. (Middle and right) shh is expressed in the epidermis adjacent to the blastema in the regenerating fin (arrowheads), and in a similarly restricted epidermal domain at the distal tips of the rays in the uninjured fin (arrowheads). Nuclei are labeled with DAPI (blue). (F) In situ hybridization of tissue sections for mkp3 and msxb. (Top) mkp3 is expressed in the distal lateral epidermis and distal-most mesenchyme of uninjured fins (arrowheads indicate epidermal expression, right), a pattern similar to its expression in the basal epidermal layer and blastema during regeneration (left). (Bottom) msxb expression in the uninjured fin is predominant in distal mesenchyme (right), reminiscent of blastemal expression of msxb after amputation (left). Scale bars: 50 μm. EXPRESSION / LABELING:
|
|
Homeostatic responses of cell proliferation and gene expression in zebrafish fins. (A) A model for `priming' homeostatic regeneration through manipulation of Fgf signaling. If developmental gene expression and cell proliferation are homeostatic events that rely on Fgf signaling, then these events should increase in intensity as a response after fins recover from a period of Fgfr inhibition. (B) BrdU incorporation in uninjured fins after a priming protocol of 14 days of daily heatshocks, and 5 days at room temperature (14d hs/5 dr). This protocol was predicted to repress, and then release and increase, homeostatic proliferation in hsp70:dn-fgfr1 fins. Transgenic fins display a burst of BrdU incorporation in distal fin tissue during recovery (brackets) that is not detectable in wild-type fins. One lobe of the caudal fin is shown. (C) Expression of regeneration marker genes increases during recovery of Fgf signaling. mkp3 and msxb are robustly expressed in regenerating fins (4 dpa, arrowheads), but expression in the uninjured fin or primed wild-type fin is undetectable by whole-mount in situ hybridization. However, mkp3 and msxb levels increase visibly following homeostatic priming of the fin by transient Fgfr inhibition (arrowheads). EXPRESSION / LABELING:
|
|
Fgf20a and Mps1 are required for homeostatic regeneration in zebrafish fins. (A,B) In situ hybridization for fgf20a and mps1 during regeneration and priming. fgf20a and mps1 expression is increased upon recovery of Fgf signaling (arrowheads), as described for mkp3 and msxb. Expression is undetectable by this method in wild-type fins. (C) Images of wild-type, fgf20a and mps1 mutant fins at day 0, day 30 and day 60 at the restrictive temperature (33°C). Both mutants exhibited a significant loss in distal tissue that was not seen in wild-type controls maintained at the restrictive temperature. (D) Quantification of length changes in centrally located rays of fgf20a and mps1 mutants. Both mutant strains showed a significant reduction in fin length after 30 and 60 days at the restrictive temperature, whereas wild-type controls maintained fin length (mean±s.e.m., Student's t-test, *P<0.001 at days 30 and 60). |
|
Fins recover lost structures after restoration of Fgf signaling. The same hsp70:dn-fgfr1 fin pictured at day 60 of Fgfr inhibition (left), and after 30 days of recovery at room temperature (right). The majority of fin rays recover ostensibly all lost structures. Inset shows region from which new growth was initiated after Fgf signaling. Arrowheads indicate the start of new tissue. PHENOTYPE:
|
|
Injuries to distal fin structures are rare under laboratory conditions. In animals imaged every 2-3 days over 14 to 24 days, we found that injuries and loss of distal bone were rare (2 events in 13 animals), and were rapidly regenerated (arrows). Scale bar: 100 μm. |
|
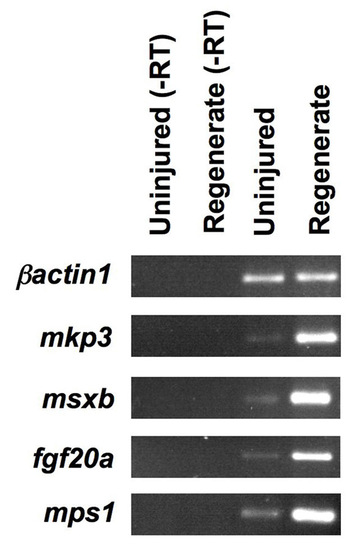
Markers of facultative regeneration are expressed at low levels in uninjured fins. The blastemal regulators mkp3, msxb, fgf20a and mps1 are all robustly expressed in samples from regenerating fins, as assessed by RT-PCR. Expression of these factors is reduced, but detectable, in the uninjured fin. The control, β-actin1, is found at similar levels in samples taken from both uninjured and regenerating fins. EXPRESSION / LABELING:
|