- Title
-
Transcription of fgf8 is regulated by activating and repressive cis-elements at the midbrain-hindbrain boundary in zebrafish embryos
- Authors
- Inoue, F., Parvin, M.S., and Yamasu, K.
- Source
- Full text @ Dev. Biol.
|
Regulatory function of the S4.2 enhancer in zebrafish embryos. (A) Schematic view of the zebrafish fgf8 gene. The five exons constituting zebrafish fgf8 (exons 1a, 1b, 1d, 2, and 3) and seven highly conserved sequences in the vicinity of fgf8 are indicated by blue and red boxes, respectively (Inoue et al., 2006). The transcriptional start site is marked with a bent arrow. The downstream gene, hagoromo (hag), is also called fbxw4. (B–D) Tg embryos showing stable expression of S4.2-EGFP or S4.2R-EGFP at 26 hpf (B, C) and 48 hpf (D). (E, G–I) Endogenous mRNA expression of fgf8 at the bud (E), 8-somite (G), 20-somite stages (H), and 34 hpf (I). (F, J–L) mRNA expression of S4.2-EGFP as determined using WMISH. (M, M′, N) Confocal microscopic views of embryos at the 16-somite stage (M, bright field; M′, epifluorescence), and a merged view at 24 hpf (N), showing GFP expression in the anteriormost hindbrain. (O, O′) Comparison of the mRNA expression of S4.2-EGFP with that of wnt1 using two-color WMISH (O, bright field; O′, epifluorescence view of the same embryo); egfp expression (purple) occurs caudal to the expression of wnt1 (red). (B–L, O, O′) Lateral views with anterior to the left and dorsal to the top (B–D, G–L, O, O′), or with anterior to the top and dorsal to the right (E, F). (M, M′, N) Dorsal views with anterior to the top. Arrowheads mark the MHB. dos, distal optic stalk; dd, dorsal diencephalon; dt, dorsal telencephalon; op, olfactory placode; os, optic stalk; ov, otic vesicle; r1/r2/r4, rhombomeres 1/2/4. Scale bars, 200 μm (B–L, J, O, O′) or 50 μm (M, M′, N). |
|
The S4.2 enhancer is composed of activating and repressive elements. (A) Sequences of the #2/#2* region and derived fragments that were examined for the regulatory activity. PBSI and the 28-bp repressive region are indicated by red letters. (B–G) Stable (B, D–G) or transient (C) GFP expression of the deleted versions of S4.2-EGFP. (H–J, H′–J′) mRNA expression of S4.2Δ2c-EGFP was visualized using WMISH. Arrowheads indicate GFP expression in the MHB. Lateral (B–J) and dorsal (H′–J′) views are shown. d, diencephalon; dos, distal optic stalk; h, hindbrain; m, midbrain; op, olfactory placode; os, optic stalk; ov, otic vesicle, r1/3/5, rhombomere 1/3/5. Scale bars, 200 μm. |
|
Pax2a is necessary and sufficient for the enhancer activity of S4.2. (A) S4.2-EGFP expression was not detected in the MHB and distal optic stalk, whereas it is retained in the olfactory placode and otic vesicle in noi embryos. (B) S4.2-EGFP expression was lost only in the MHB, leaving GFP expression in the sensory regions intact in ace embryos. (C–G) fgf8 expression in the heads of noi embryos (C), ace embryos (D, G), and wild-type (WT) embryos (E, F) was examined by WMISH at 26 hpf. The insets in panels C–E show fgf8 expression in the otic vesicles. Black and white arrows show the presence and absence, respectively, of reporter expression in the distal optic stalks. (H, I) Misexpression of pax2a by mRNA injection caused ectopic induction of endogenous fgf8 in wild-type embryos (H) and egfp in S4.2-EGFP Tg embryos (I), as are marked with white arrowheads. (J, J′) S4.2-EGFP Tg embryos were injected with S4.2Δ2c-pax2a DNA and subjected to two-color in situ analysis at the 16-somite stage; first for pax2a (J, J′) and additionally for egfp (J′). Targeted misexpression of pax2a led to ectopic induction of egfp (purple) within the pax2a-positive domain (red, small white arrowheads) in the hindbrain. Black arrowheads indicate the MHB. Lateral views (A–E, H, I), frontal views (F, G), or dorsal views (J, J′) are shown. dd, dorsal diencephalon; dos, dorsal optic stalk; op, olfactory placode; os, optic stalk; ov, otic vesicle; pos, proximal optic stalk; t, telencephalon. Scale bars, 200 μm (A–I) or 100 μm (J, J′). EXPRESSION / LABELING:
|
|
Binding of Pax2a to the S4.2 enhancer in vitro and in vivo. (A) Subfragments of the S4.2 region examined using EMSA in panels C–E. See Fig. 5A for sequences #2a, #2b*, and #2c*. (B) Alignment of the sequences of the oligos for PBSI (PI), PBSII (PII), and BS-I (Song et al., 1996) with the consensus sequences for Pax2 (Epstein et al., 1994) and Pax5 (Czerny et al., 1993). Conserved nucleotides are underlined. Base substitutions in the mutated forms of PI and PII (PIm and PIIm) are indicated in lowercase. (C) Binding of Pax2a with the reference probe, BS-I, was challenged using a 100-fold molar excess of the respective subfragments as competitors. (D) Direct binding of PBSI and PBSII to Pax2a and Pax8 using DIG-labeled PI and PII probes. (E) Binding of Pax2a to DIG-labeled PI and PII was competed efficiently by a 100-fold molar excess of the corresponding DNA, but their competing activity was abrogated by base substitutions within the core binding sequences (PIm and PIIm). (F) The regions covering PBSI and PBSII were amplified separately from immunoprecipitates obtained by treating the sonicated chromatin of 16-somite-stage embryos with anti-Pax2 (ChIP assay); no amplification occurred when the chromatin was incubated with or without IgG. |
|
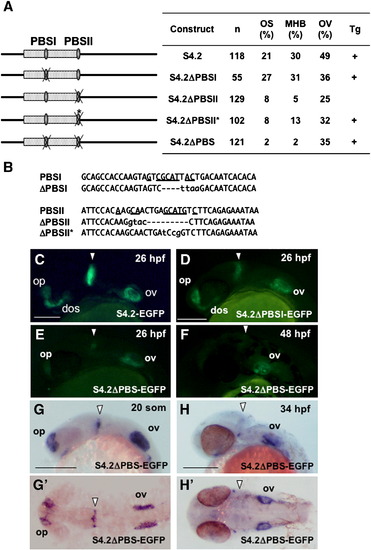
Essential role of the two Pax2 binding sites in the regulatory function of the S4.2 enhancer. (A) Disruption of the Pax2 binding sites (PBSI and PBSII) in S4.2-EGFP. The percentages of injected embryos expressing GFP in the optic stalk (OS), MHB, and the otic vesicle (OV) at 24–26 hpf relative to the numbers of GFP-positive embryos (n) are shown on the right. The constructs for which expression was confirmed by stable expression in Tg embryos are marked with (+). (B) Mutations introduced into the two Pax2-biniding sites of S4.2-EGFP. Deletions and base substitutions are indicated by hyphens and lowercase letters, respectively. In case of ΔPBSI and ΔPBSII, an EcoRI and a KpnI sequence were introduced into the Pax2-binding core sites by inverse PCR, respectively, resulting in base deletion and substitution. Meanwhile, the Pax2 site was disrupted by site-directed mutagenesis for ΔPBSII*. (C–F) Stable GFP expression in Tg embryos harboring the intact (C) or mutated S4.2-EGFP constructs (D–F) was observed at 26 hpf (C–E) and 48 hpf (F). (G, H, G′, H′) Expression of egfp mRNA in the head of an S4.2ΔPBS-EGFP Tg embryo. Expression of S4.2ΔPBS-EGFP in the MHB was weak relative to that in the sensory regions at 19 hpf (20-somite stage)–26 hpf (E, G, G′), and it was disrupted later (F, H, H′). Lateral (C–H) and dorsal (G′, H′) views are shown with anterior to the left. Arrowheads indicate the MHB. dos, distal optic stalk; op, olfactory placode; ov, otic vesicle. Scale bars, 200 μm. EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 316(2), Inoue, F., Parvin, M.S., and Yamasu, K., Transcription of fgf8 is regulated by activating and repressive cis-elements at the midbrain-hindbrain boundary in zebrafish embryos, 471-486, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.