- Title
-
RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA
- Authors
- Kedde, M., Strasser, M.J., Boldajipour, B., Oude Vrielink, J.A., Slanchev, K., le Sage, C., Nagel, R., Voorhoeve, P.M., van Duijse, J., Andersson Ørom, U., Lund, A.H., Perrakis, A., Raz, E., and Agami, R.
- Source
- Full text @ Cell
|
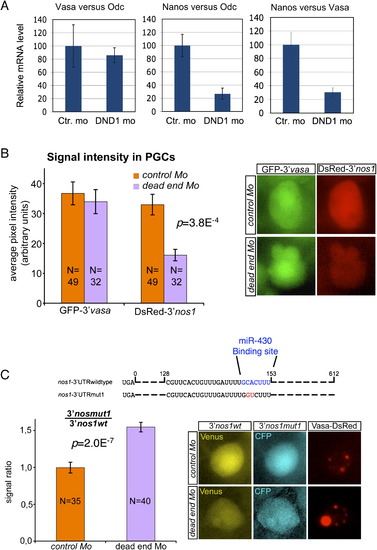
Zebrafish Dnd1 (drDnd1) Counteracts Inhibition of Nanos by miR-430 |
|
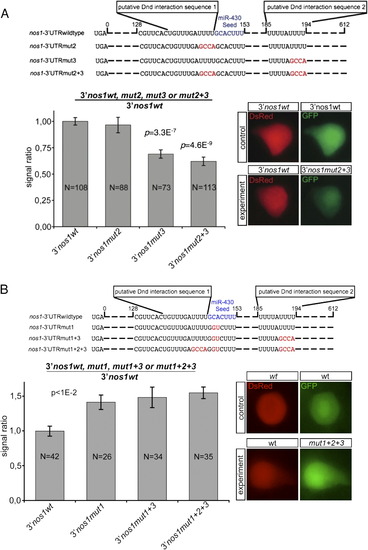
URRs Are Required for Dnd1 to Efficiently Repress miR-430-Mediated Nanos Inhibition |
|
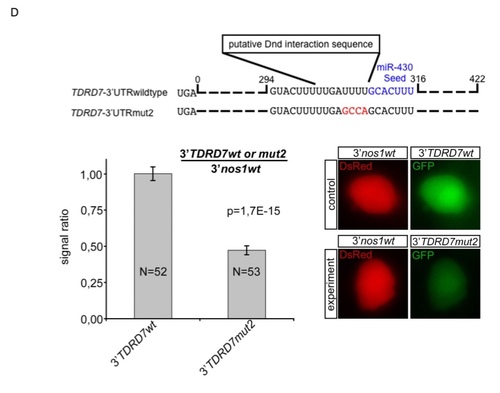
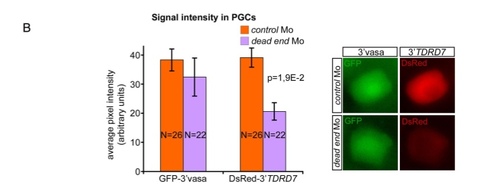
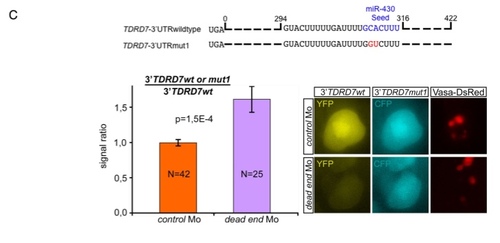
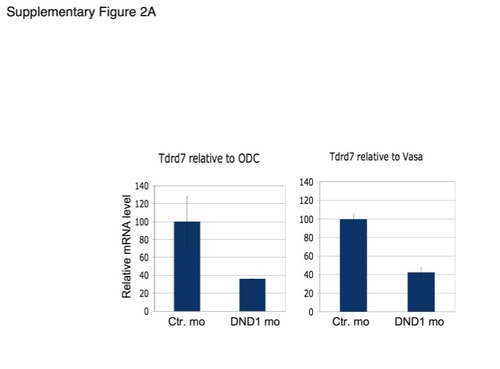
Zebrafish DND1 (drDND1) counteracts inhibition of TDRD7 by miR-430 through binding to adjacent URRs. |
Reprinted from Cell, 131(7), Kedde, M., Strasser, M.J., Boldajipour, B., Oude Vrielink, J.A., Slanchev, K., le Sage, C., Nagel, R., Voorhoeve, P.M., van Duijse, J., Andersson Ørom, U., Lund, A.H., Perrakis, A., Raz, E., and Agami, R., RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA, 1273-1286, Copyright (2007) with permission from Elsevier. Full text @ Cell