- Title
-
Polysialyltransferase expression is linked to neuronal migration in the developing and adult zebrafish
- Authors
- Rieger, S., Volkmann, K., and Köster, R.W.
- Source
- Full text @ Dev. Dyn.
|
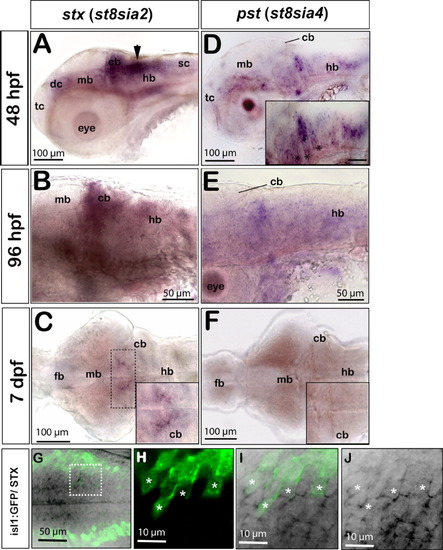
stx and pst expression in the differentiating zebrafish central nervous system. Panels A,B,D,E show lateral and C,F dorsal views, anterior is to the left. A: At 48 hours postfertilization (hpf) stx expression is weakly present in the ventral telencephalon, in the diencephalon, midbrain, in the floorplate, and in the spinal cord. Strong expression is found in dorsal regions of the caudal hindbrain along the fourth ventricle (black arrow) and in the differentiating cerebellum. B: stx expression remains weakly detectable in the differentiating cerebellum and in ventral regions of the mid- and hindbrain at 96 hpf. C: At 7 days postfertilization (dpf), only the dorsomedial domain of the hindbrain retains low levels of stx (dashed box and inset showing high magnification of the dorsomedial domain in the anterior hindbrain with remaining stx expression). D: At 48 hpf, pst is expressed in the ventral domains of the fore- and midbrain. A striped expression pattern can be observed in the hindbrain, being confined to the central domains of individual rhombomeres (inset, asterisks). E: Only some patches in the hindbrain retain low pst expression levels at 96 hpf. F: pst expression is no longer detectable in the brain at 7 dpf (compare inset in F with inset in C). G-J: Expression of stx in a subset of cranial motoneurons as revealed by colabeling of stx and green fluorescent protein (GFP) in embryos of the islet1:GFP transgenic line. G: Maximum intensity projection of a 30-μm stack of optical sections showing stx expression in medial domains of the hindbrain (black) and more laterally positioned GFP-expressing cranial motoneurons (green). H-J: A subset of cranial motoneurons (H, white asterisks) expresses stx (J, white asterisks), which can be identified in 1-μm single optical sections at high magnification. I: Overlay of GFP and stx expression. cb, cerebellum; dc, diencephalon; fb, forebrain; hb, hindbrain; mb, midbrain; sc, spinal cord; tc, telencephalon. |
|
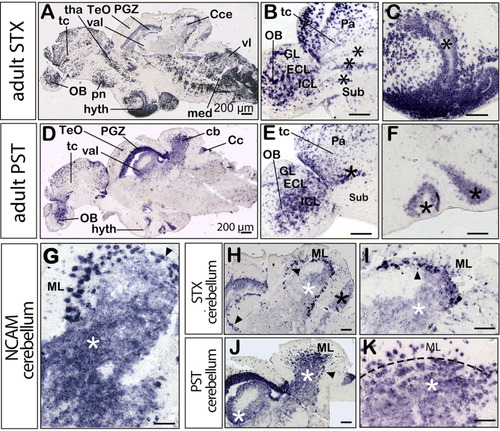
Comparison of stx and pst expression in the adult zebrafish brain. All panels show medial sagittal sections, anterior is to the left. A-C,H,I show stx expression and D-F,J,K show pst expression. A: Overview of stx expression. D: Overview of pst expression. B,E: Cells in the pallial migratory domains of the telencephalon and olfactory bulb are marked by asterisks. C,F: Ventricular staining in the hypothalamic region. G: ncam expression in the molecular layer (ML), Purkinje cell layer (PCL, black arrowhead), and granule cell layer (GCL, white asterisk) of the cerebellum. H: Higher magnification of the cerebellum showing stx expression in some cells of the ML and strong expression in cells of the PCL, extending from the corpus into the valvula cerebelli (black arrowheads). Weak expression levels are present in the caudal lobe (black asterisk) and in the GCL (white asterisk). I: A higher magnification of stx expression in the adult cerebellum. J: Strong pst expression in the GCL of the corpus cerebelli and in the valvula cerebelli being more pronounced than stx expression (H, white asterisk). K: A higher magnification of pst expression in the adult cerebellum, being detected in the ML and GCL, separated by a dashed line. Cc, crista cerebellaris; Cce, corpus cerebelli; ECL, external cellular layer; GL, glomerular layer; hyth, hypothalamus; IL, internal cellular layer; med, medulla; ML, molecular layer; OB, olfactory bulb; Pa, pallium; PGZ, periventricular gray zone of the optic tectum (TeO); pn, preoptic nucleus; Sub, subpallium; tc, telencephalon; tha, thalamus; val, valvula cerebelli; vl, vagal lobe. Scale bars = 200 μm. EXPRESSION / LABELING:
|
|
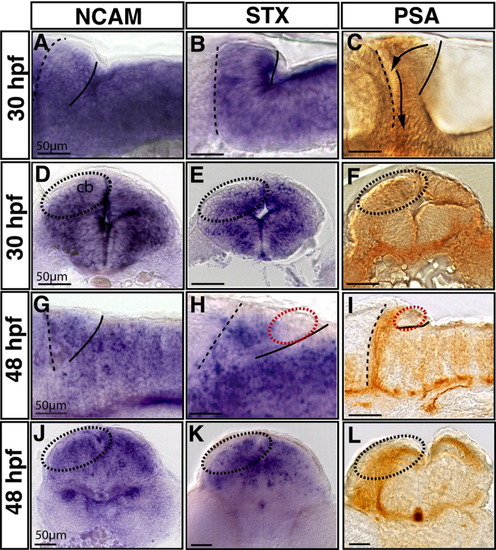
Comparison of ncam, stx and polysialic acid (PSA) expression in the differentiating cerebellum during stages of neuronal migration. A-L: High magnification of lateral (A-C,G-I) and transverse (D-F,J-L) sections of the anterior hindbrain are shown at 30 hpf (A-F) and 48 hpf (G-L). The anterior cerebellar border at the midbrain-hindbrain boundary (MHB) and the posterior edge of the cerebellum are marked with a dashed and solid line, respectively. The mRNA in situ analysis was performed for ncam (A,D,G,J) and stx expression (B,E,H,K), while PSA immunoreactivity was detected using the mAb735 antibody (C,F,I,L). At 30 hours postfertilization (hpf), ncam expression was found to be strong and uniform in the cerebellum (A,D) similar to stx expression showing strongest expression at the ventricle (B,E). Moderate PSA levels can also be detected in this domain (C,F) during onset of rhombic lip derived neuronal migration (compare areas of dashed oval circles demarcating one cerebellar half). G,J: At 48 hpf, ncam expression is still uniform in the cerebellum. H,I: In contrast, stx and PSA expression have become restricted to the rostral cerebellar region along the MHB and are not longer detectable along the ventricle (red dashed circle). K,L: Transverse views of stx and PSA expression in the anterior cerebellum delineating the anteroventral migration route of neuronal progenitors. Scale bars = 50 μm. |
|
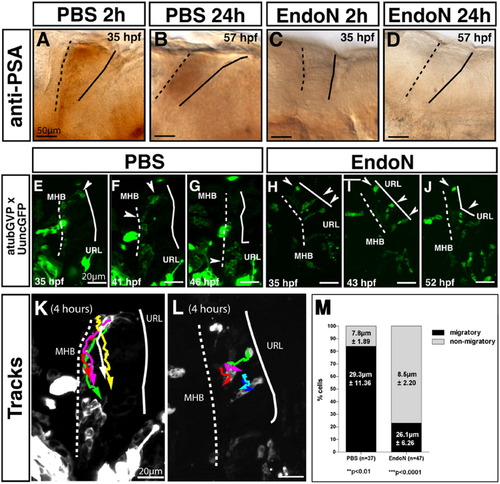
Polysialic acid (PSA) degradation in the central nervous system of EndoN-treated embryos impairs migration from the cerebellar rhombic lip. All panels show lateral views, anterior is to the left. Panels A-D show PSA expression in cerebellar primordia after phosphate buffered saline (PBS) and EndoN treatment at 33 hours postfertilization (hpf). A,B: Control embryos injected with PBS show PSA immunoreactivity after 2 hr (A) and 24 hr (B). Note the weaker expression levels at 57 hpf are due to endogenous down-regulation of PSA. C,D: EndoN injection at 33 hpf completely abolished PSA expression already after 2 hr (C), lasting up to 24 hr (D). Panels E-J are maximum intensity projections (50-μm stacks recorded every 12 min) showing migrating neuronal progenitors in the differentiating cerebellum after PBS (E-G) and EndoN treatment (H-J), recorded by in vivo time-lapse confocal microscopy. E: A group of neuronal progenitor cells at the upper rhombic lip (URL) in the control embryo is marked by a white arrowhead 2 hr post-injection. F: These cells migrated approximately halfway between URL and midbrain-hindbrain boundary (MHB) after 6 hr, while others have reached the MHB (white arrowhead). G: After 11 hr, cells in the ventral cerebellar region become stationary and form a cluster (white arrowhead, see Supplementary Movie S1). H: After injection of EndoN into the hindbrain ventricle, neuronal progenitors marked by an arrowhead are positioned at the URL or are polarized and span across the cerebellum between the URL and MHB (white arrowheads). I,J: Eight hours later (I), these neuronal progenitor cells are stalled at the URL and remain in this position (J) for up to 17 hr of time-lapse recording (see Supplementary Movie S2). K,L: The migratory routes of five individual uncGFP-expressing neuronal progenitor cells in PBS (K) and EndoN (L) -treated embryos were traced using the manual tracking tool plug-in of ImageJ1.34, the migratory direction is marked by an arrowhead. K: Anteroventral migration of cells in a PBS-injected embryo. L: Reduced migration of cells in EndoN-injected embryo. M: Quantification of neuronal progenitor migration from the cerebellar rhombic lip in PBS- and EndoN-treated embryos. While 84% of neuronal progenitors (n = 5 movies, 37 cells analyzed) in PBS-treated embryos were migratory, covering an average distance of almost 30μm during a 4-hr period, only 23% of neuronal progenitors (n = 5 movies, 47 cells analyzed) in EndoN-treated embryos were migratory and covered a similar average migration distance. Cells were classified as nonmigratory when they moved less than one cell diameter (approximately 10μm) during the observation period. |

Unillustrated author statements EXPRESSION / LABELING:
|