- Title
-
Early Endocardial Morphogenesis Requires Scl/Tal1
- Authors
- Bussmann, J., Bakkers, J., and Schulte-Merker, S.
- Source
- Full text @ PLoS Genet.
|
A Truncating Mutation in the Zebrafish tal1 Gene (A) Live micrographs of wt and tal1t21384 mutant embryos at 56h. Mutant embryos have slightly curved tails, smaller eyes, and pericardial edema. (B) Electropherograms of wt (+/+), heterozygous (+/-) and tal1t21384 homozygous genomic DNA from bp 7–17 of the third tal1 coding exon. The A→T transversion (arrow) mutates lysine 183 to an ochre (TAA) stop codon (K183X). (C) Schematic diagram of the primary Tal1 protein structure. The DNA-binding bHLH domain is indicated in yellow. The tal1t21384 mutation deletes the Tal1 protein C terminus including the complete helix-loop-helix domain (D) The tal1t21384 mutation leads to severe defects in intersomitic vessel formation at 36 hpf, visualized in a fli1a:gfp transgenic background. Early endothelial pattering can be rescued by injecting 20 pg wt tal1 mRNA into mutant embryos. (E) Loss of tal1 expression in tal1t21384 mutant embryos. In situ hybridization for tal1 shows a loss of tal1 expression in erythroid cells and a reduction of tal1 expression in the spinal chord. tal1 expression is retained at normal levels in some hematopoietic/endothelial progenitor cells in the tail (arrowheads) and in some mesenchymal cells of unknown nature just dorsal to the yolk extension (arrows). These bilateral cell populations lie ventral to the pronephric tubules and lateral to the gut endoderm (insets). Also note the loss of tal1 expression in the ventral wall of the dorsal aorta containing, in wt embryos, the definitive hematopoietic stem cells. (F) Loss of hematopoiesis in tal1t21384 mutant embryos. In situ hybridization for hematopoietic markers at 26 hpf shows a loss of primitive erythroid cells (gata1), primitive myeloid cells (pu.1), and definitive hematopoietic stem cells (runx1). |
|
Defects in Endocardial Development in tal1t21384 Mutants (A) Confocal images of the head region from embryos transgenic for fli1a:gfp at 32 hpf. Arrows indicate the location of the endocardium. In the wt endocardium, a lumen is visible, which is absent in tal1t21384 mutants. (B) Transversal sections of wt and tal1t21384 mutant heart ventricles at 56 hpf stained with hematoxylin/eosin. In wt embryos, the endocardium forms an epithelium attached to the myocardium, and blood cells are visible within the heart tube. In the tal121384 mutant heart, the endocardium fails to form an epithelium and is only present in the heart ventricle. (C) Rescue of endocardial defects. Maximum z-projection of a stack of confocal images of embryos carrying the vegfr4:gfp transgene. Embryos were divided into three classes based on their endocardial phenotype. Class 1: wt length of the endocardium with normal tube formation (wt phenotype). Class 2: wt length of the endocardium but no tube formation (intermediate phenotype). Class 3: short endocardium with no tube formed (mutant phenotype). All embryos homozygous or heterozygous for the wt allele are in class 1, whereas all embryos homozygous for the tal1t21384 allele are in class 3, showing the high penetrance of this phenotype. In 43 out of 54 embryos homozygous for the tal1t21384 allele (83%), the endocardial phenotype can be rescued to class 1 through injection of low amounts of wt tal1 mRNA (20 pg per embryo), whereas most of the remaining (8/54, 15%) display an intermediate phenotype (class 2). PHENOTYPE:
|
|
Zebrafish vegfr4(flk1) Represents a New Class of Nonmammalian VEGF Receptors (A) Rooted neighbor-joining tree of vertebrate VEGF receptors. Different colors represent different classes of VEGF receptors. Note the clear separation of zebrafish and other teleost fish vegfr4, and the novel frog, chick, opossum, and platypus VEGF receptor genes from the three other classes of vertebrate VEGF receptors. Bootstrap value of VEGFR4 node is indicated (1,000 times out of 1,000 iterations). (B) Synteny analysis of vertebrate VEGF receptors using human, mouse, chick, and zebrafish genome assemblies. Colors used are similar to those in (A). Question marks represent missing orthologous genes, potentially due to gaps in the chick genome assembly. Clear syntenic relationships of all vertebrate VEGF receptors were observed, indicating duplication from a primordial gene cluster in a primitive chordate. Mammals have lost the Zebrafish vegfr4 orthologue, coinciding with the emergence of the X-chromosome inactivation center XIST in the same genomic locus. (C) Expression of zebrafish VEGF receptors in the heart region at 26 hpf, dorsal view, anterior to the top. Three VEGF receptors are expressed in the endocardium: vegfr4, kdr, and flt1 (arrowheads). Note the absence of flt4 expression in the endocardium (asterisk). At this stage, both vegfr4 and kdr are expressed at high levels in all endothelial cells, whereas flt1 and flt4 have a mutually exclusive expression pattern: flt1 is expressed in all arteries (AA1, first aortic arch; LDA, lateral dorsal aorta; and PICA, primitive internal carotid artery) whereas flt4 is expressed in all veins (MCeV, middle cerebral vein [which is largely out of the focal plane of this picture]). PHBC, primordial hindbrain channel; PMBC, primordial midbrain channel. EXPRESSION / LABELING:
|
|
Migration of Endocardial Precursors in wt Embryos (A) Embryos transgenic for vegfr4:gfp were subjected to time-lapse confocal microscopy, revealing rapid endocardial migration prior to heart tube formation. A movie demonstrating this process can be viewed in the Video S1. Twelve individual frames from this movie at indicated stages and time-points are shown seen in (A), with white dashed lines indicating the position of (pre-) endocardial cells. Frames 1–5 show the fusion of the bilateral endocardial precursors between the 14- and 16-somite stages. Frames 5–8 indicate the posterior migration of endocardial cells to cover the lateral and posterior regions of the cardiac disc; note the posterior migration of the paired lateral dorsal aortas between the 18- and 22-somite stages (white arrows). The apex of the endocardial disc (pink dashed line) appears to be constricted below the aortic arches between the 18- and 22-somite stages (frames 7–10). A leftward movement of the endocardium is visible between the 20- and 26-somite stages (frames 9–12), and is coinciding with the appearance of single vegfr4:gfp-positive cells lateral to the remaining endocardium (red dashed line). Also note the migration of the venous posterior hindbrain channels (red arrows) between the 22- and 26-somite stage (frames 10–12). (B) Relative locations of endocardial and myocardial precursors during fusion of endocardial precursor populations, revealed by two-color in situ hybridization showing cdh5 (blue, endocardium) and nkx2.5 (red, myocardium) expression. The bilateral populations of endocardial precursors (arrows) are located anterior to the myocardial precursors until the 14-somite stage, then migrate medially and posteriorly to assume a position in between the myocardial precursors at the 18-somite stage. (C) Embryos transgenic for fli1a:gfp were subjected to time-lapse confocal microscopy, revealing slow medial movement of gfp-positive cells between the six- and 12-somite stage (frames 1–4) and rapid migration starting at the 14-somite stage. A movie demonstrating this process can be viewed in the Video S2. EXPRESSION / LABELING:
|
|
Migration of Endocardial Precursors and Heart Tube Formation in wt tal1t21384 Mutant Embryos (A) tal121384 mutant embryos transgenic for vegfr4:gfp were subjected to time-lapse confocal microscopy, revealing defects during early endocardial precursor migration. A movie demonstrating this process can be viewed in the Video S3. Six individual frames from this movie are shown in (A). Whereas the initial formation of bilateral endocardial precursors is not affected, posterior migration is disturbed, and endocardial precursors remain attached in a relative anterior position. Note that migration of the paired lateral dorsal aortae (arrows) proceeds normally. (B) tal1 expression in endocardial precursors. Two-color in situ hybridization revealing tal1 (blue) and nkx2.5 (red) expression in wt embryos at the 14- and 18-somite stage. tal1 expression is observed in endocardial but not myocardial precursors during their posterior migration (arrowheads). (C) Expression of the arterial markers flt1 and hey2 is retained in tal1t21384 mutant endocardium, but severely reduced in endothelium, as revealed by in situ hybridization. Dorsal view of flt1 and hey2 expression in the head, anterior to the top, and lateral view of flt1 and hey2 in the tail (28 hpf). In wt embryos, flt1 expression is observed in all head arteries, the aortic arches and the endocardium (arrow). In tal1t21384 mutant embryos, expression of flt1 is observed in a few remaining head arteries and the aortic arches. High levels are seen in the endocardium (arrow). In wt embryos, flt1 expression is observed in the dorsal aorta and the developing intersegmental vessels. In the tail of tal1t21384 mutant embryos, expression of flt1 is abolished, except for a few remaining cells that express flt1 at low levels (arrowheads). In wt embryos, hey2 expression is observed in the endocardium and the aortic arches and in some parts of the brain and spinal chord. In tal1t21384 mutant embryos, expression in the endocardium (arrow) is increased. In wt embryos, hey2 expression is observed in the dorsal aorta and the developing intersegmental vessels, spinal chord neurons, and in ventral and dorsal cells of the somites. In tal1t21384mutant embryos, expression in the dorsal aorta and intersegmental vessels is severely reduced, although some anterior intersegmental vessels and aortic cells retain low levels of hey2 expression (arrowheads). EXPRESSION / LABELING:
|
|
Defective Heart Tube Formation in tal1t21384 Mutant Embryos Despite Normal Fusion of Bilateral Myocardial Precursor Populations (A) Chamber differentiation in wt and tal1t21384 mutant embryos revealed by two-color in situ hybridization showing amhc (atrium) and cmlc2 (atrium and ventricle) at 28 hpf. Chamber differentiation proceeds normal in mutant embryos, but the primary heart tube is malformed with an enlarged atrial inflow region (brackets). (B–D). Two-color immunohistochemistry using anti-tropomyosin (red, myocardium) and anti-gfp (green, vegfr4, endocardium) antibodies. Images were generated as maximum projections of confocal z-stacks (ventral views, anterior to the top). Some yolk platelets show intense autofluorescence at the wavelength used for anti-tropomyosin detection (647 nm). (B) Heart morphogenesis during heart field fusion in wt embryos. The bilateral heart fields fuse dorsal to the endocardium between the 18- and 20-somite stage. Fusion is initiated in the posterior region of the heart. At the 18-somite stage, tropomyosin-positive cells are located ventral to the first aortic arches. (C) Endocardial precursors are ventral to the myocardium in the lateral and posterior regions of the heart. Endocardial and myocardial sheets are closely associated, as relative positions were only revealed after deconvolution of confocal stacks. Four deconvolved images of the confocal image stack in (B) are shown in (C). (D) In tal1t21384 mutant embryos, initial fusion of the myocardial precursor populations occurs normally, although endocardial precursors are absent in the posterior region of the heart field. (E) Primary heart tube formation from myocardial and endocardial precursors in wt and tal1t21384 mutant embryos revealed by two-color in situ hybridization showing cdh5 (endocardium, blue) and cmlc2 (myocardium, red) expression. Dorsal views, anterior is to the top. At the 22-somite stage, wt embryos have formed a cardiac disc, with endocardial cells underlying the circular myocardial primordium. The medial endocardium within the ring of myocardium forms the connection between the endocardium and the aortic arches. In tal1t21384 mutant embryos, anterior closure of the myocardial primordium is defective due to aggregation of endocardial precursors. At the 26-somite stage, wt embryos have formed the primary heart tube and rhythmical contractions begin. In tal1t21384 mutant embryos, heart tube formation is delayed. By this stage, myocardial cells have completed fusion formation at the anterior side of the cardiac disc. (F) Schematic overview of fusion of myocardial precursor regions and heart tube formation in wt and tal1t21384 mutant embryos. Endocardium and aortic arches are in green, myocardium in red. EXPRESSION / LABELING:
PHENOTYPE:
|
|
kdr Expression during Gastrulation. At the tailbud (TB) stage, kdr but not vegfr4 is expressed. Expression of kdr is diffuse, but highest around the tailbud and near the embryonic axis. |
|
Endothelial Gene Expression in the Trunk Trunk expression of different markers in wt and tal1t21384 mutant embryos at 24–30 hpf for general endothelial (cdh5, vegfr4), venous (flt4, dab2), and arterial (notch3, ephrinB2) differentiation. In mutant embryos, expression of arterial marker genes is reduced or absent, whereas the expression domain of venous marker genes is expanded to the dorsal aorta. |
|
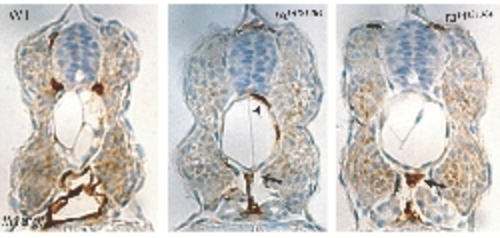
Presence of Endothelial Cells in the Region of the Dorsal Aorta Immunohistochemistry using anti-GFP antibody in the fli1a:gfp background; brown labeling indicates gfp-positive endothelial cells. Although no luminized vessels in the trunk are formed in tal1t21384 mutants, endothelial cells are present in the location of the dorsal aorta (arrows), and some intersegmental vessels form (arrowhead). PHENOTYPE:
|

Unillustrated author statements |